Multiple regression model - numeric covariates
Jan Vávra
Exercise 4
Download this R markdown as: R, Rmd.
Outline of the lab session
- Multiple regression model for the continuous response vector with continuous regressors
- K-variate parametrizations of a numeric covariate
Loading the data and libraries
The dataset mtcars (available under the standard R
installation) will be used for the purposes of this lab session. Some
insight about the data can be taken from the help session (command
?mtcars) or from some outputs provided below.
library(colorspace)
head(mtcars)## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
## Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
## Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
## Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1str(mtcars)## 'data.frame': 32 obs. of 11 variables:
## $ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
## $ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
## $ disp: num 160 160 108 258 360 ...
## $ hp : num 110 110 93 110 175 105 245 62 95 123 ...
## $ drat: num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
## $ wt : num 2.62 2.88 2.32 3.21 3.44 ...
## $ qsec: num 16.5 17 18.6 19.4 17 ...
## $ vs : num 0 0 1 1 0 1 0 1 1 1 ...
## $ am : num 1 1 1 0 0 0 0 0 0 0 ...
## $ gear: num 4 4 4 3 3 3 3 4 4 4 ...
## $ carb: num 4 4 1 1 2 1 4 2 2 4 ...summary(mtcars)## mpg cyl disp hp drat wt qsec
## Min. :10.4 Min. :4.00 Min. : 71.1 Min. : 52.0 Min. :2.76 Min. :1.51 Min. :14.5
## 1st Qu.:15.4 1st Qu.:4.00 1st Qu.:120.8 1st Qu.: 96.5 1st Qu.:3.08 1st Qu.:2.58 1st Qu.:16.9
## Median :19.2 Median :6.00 Median :196.3 Median :123.0 Median :3.69 Median :3.33 Median :17.7
## Mean :20.1 Mean :6.19 Mean :230.7 Mean :146.7 Mean :3.60 Mean :3.22 Mean :17.8
## 3rd Qu.:22.8 3rd Qu.:8.00 3rd Qu.:326.0 3rd Qu.:180.0 3rd Qu.:3.92 3rd Qu.:3.61 3rd Qu.:18.9
## Max. :33.9 Max. :8.00 Max. :472.0 Max. :335.0 Max. :4.93 Max. :5.42 Max. :22.9
## vs am gear carb
## Min. :0.000 Min. :0.000 Min. :3.00 Min. :1.00
## 1st Qu.:0.000 1st Qu.:0.000 1st Qu.:3.00 1st Qu.:2.00
## Median :0.000 Median :0.000 Median :4.00 Median :2.00
## Mean :0.438 Mean :0.406 Mean :3.69 Mean :2.81
## 3rd Qu.:1.000 3rd Qu.:1.000 3rd Qu.:4.00 3rd Qu.:4.00
## Max. :1.000 Max. :1.000 Max. :5.00 Max. :8.001. Multiple linear regression model
For simplicity, we will only consider three explanatory (continuous)
variables from the whole mtcars dataset, i.e. \(\boldsymbol{x} \in \mathbb{R}^3\). Any
generalization for general \(\boldsymbol{X}
\in \mathbb{R}^p\) for some \(p \in
\mathbb{N}\) is straightforward. In particular, the (continuous)
dependent variable \(Y \in \mathbb{R}\)
is the car consumption (variable in the dataset denoted as
mpg – miles per gallon) and three independent variables
will be the car’s displacement (disp), horse powers
(hp), and the rear axle ratio information
(drat).
Three simple scatterplots showing the dependent variable \(Y\) against one independent variable (based on the random sample \(\{(Y_i, \boldsymbol{X}_i^\top)^\top;~i = 1, \dots, 32\}\)) are shown below. Only a limited information about the whole multivariate structure of the underlying data can be, howver, taken from the series of two-dimensional scatterplots.
par(mfrow = c(1,3), mar = c(4,4,0.5,0.5))
plot(mpg ~ disp, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue",
ylab = "Miles per gallon", xlab = "Displacement [cubic inches]")
lines(lowess(mtcars$mpg ~ mtcars$disp), col = "red", lwd = 2)
plot(mpg ~ hp, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue",
ylab = "", xlab = "Horse Power")
lines(lowess(mtcars$mpg ~ mtcars$hp), col = "red", lwd = 2)
plot(mpg ~ drat, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue",
ylab = "", xlab = "Rear axle ratio")
lines(lowess(mtcars$mpg ~ mtcars$drat), col = "red", lwd = 2)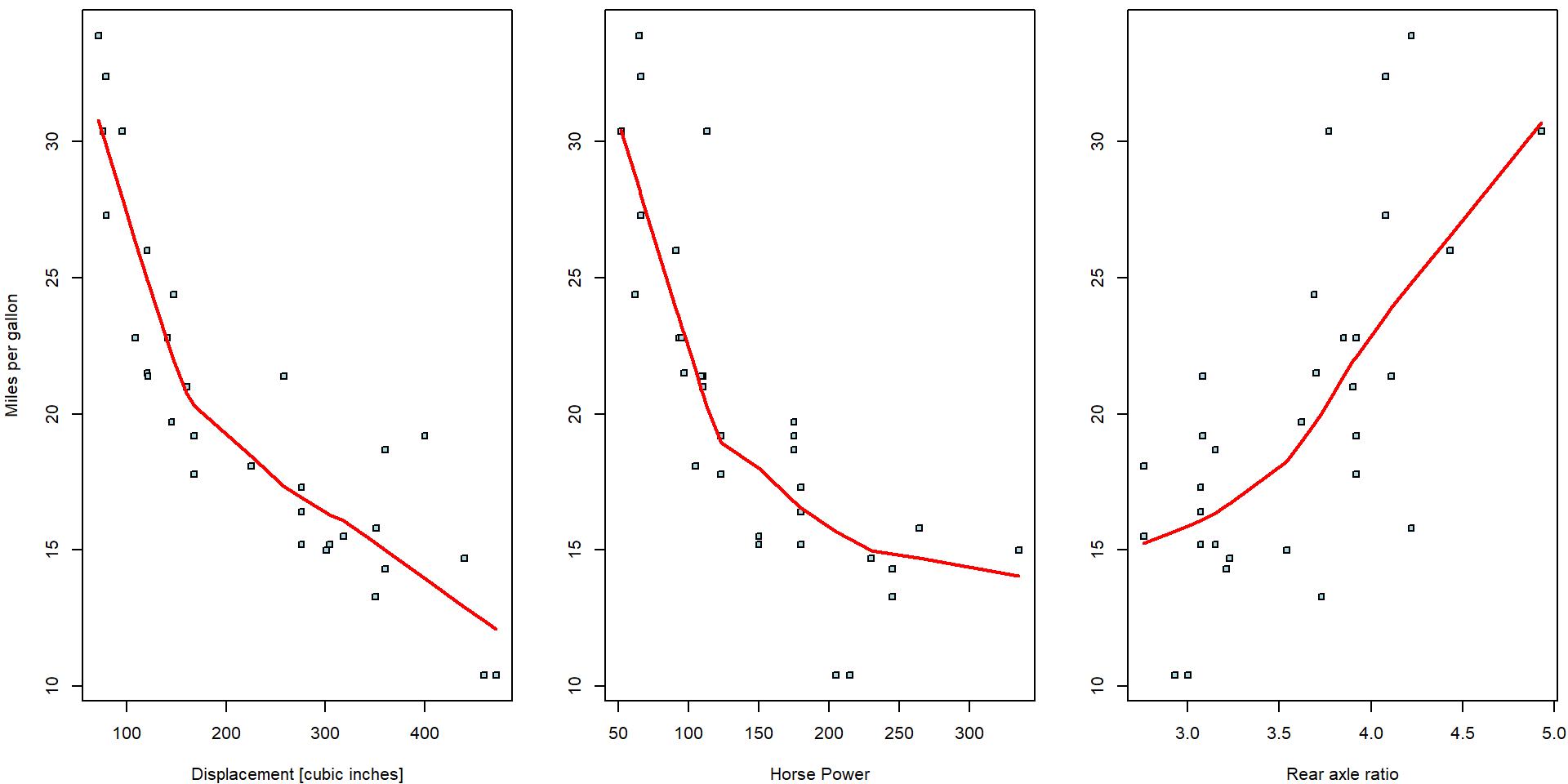
The red curves are non-parametric estimates of the conditional mean—these estimates offer a useful insight that the dependence of the consumptions is very likely negative (regressive) wrt. the displacement and horsepower while it seems to positive (progressive) for the rear axle ratio.
A simple multiple regression model of the form \[ Y = \beta_0 + \beta_1 \mathtt{disp} + \beta_2\mathtt{hp} + \beta_3 \mathtt{drat} + \varepsilon \] or mathematically (the model for the empirical data, for \(i = 1, \dots, 32\)) \[ Y_i = \boldsymbol{X}_i^\top\boldsymbol{\beta} + \varepsilon_i \] where \(\boldsymbol{X} = (1, \mathtt{disp}, \mathtt{hp}, \mathtt{drat})^\top\) and \(\boldsymbol{\beta} \in \mathbb{R}^4\) will be considered. In the R software, the model can be fitted using the following R code:
fit1 <- lm(mpg ~ disp + hp + drat, data = mtcars)
print(fit1)##
## Call:
## lm(formula = mpg ~ disp + hp + drat, data = mtcars)
##
## Coefficients:
## (Intercept) disp hp drat
## 19.3443 -0.0192 -0.0312 2.7150The information in the output above gives values for four estimated parameters – elements of the parameter vector \(\boldsymbol{\beta} \in \mathbb{R}^4\). The estimated parameters are not the same as estimated three independent simple linear regression model. Compare the output above with the following outputs:
fit_disp <- lm(mpg ~ disp, data = mtcars)
fit_hp <- lm(mpg ~ hp, data = mtcars)
fit_drat <- lm(mpg ~ drat, data = mtcars)
print(fit_disp)##
## Call:
## lm(formula = mpg ~ disp, data = mtcars)
##
## Coefficients:
## (Intercept) disp
## 29.5999 -0.0412print(fit_hp)##
## Call:
## lm(formula = mpg ~ hp, data = mtcars)
##
## Coefficients:
## (Intercept) hp
## 30.0989 -0.0682print(fit_drat)##
## Call:
## lm(formula = mpg ~ drat, data = mtcars)
##
## Coefficients:
## (Intercept) drat
## -7.52 7.68Indeed, while the signs of the estimated parameters correspond, the magnitudes are substantially different. The ordinary regressions can be directly plotted into the corresponding scatterplots:
par(mfrow = c(1,3), mar = c(4,4,0.5,0.5))
plot(mpg ~ disp, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue", ylab = "Miles per gallon", xlab = "Displacement [cubic inches]")
lines(lowess(mtcars$mpg ~ mtcars$disp), col = "red", lwd = 2)
abline(fit_disp, col = "blue")
plot(mpg ~ hp, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue", ylab = "", xlab = "Horse Power")
lines(lowess(mtcars$mpg ~ mtcars$hp), col = "red", lwd = 2)
abline(fit_hp, col = "blue")
plot(mpg ~ drat, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue", ylab = "", xlab = "Rear axle ratio")
lines(lowess(mtcars$mpg ~ mtcars$drat), col = "red", lwd = 2)
abline(fit_drat, col = "blue")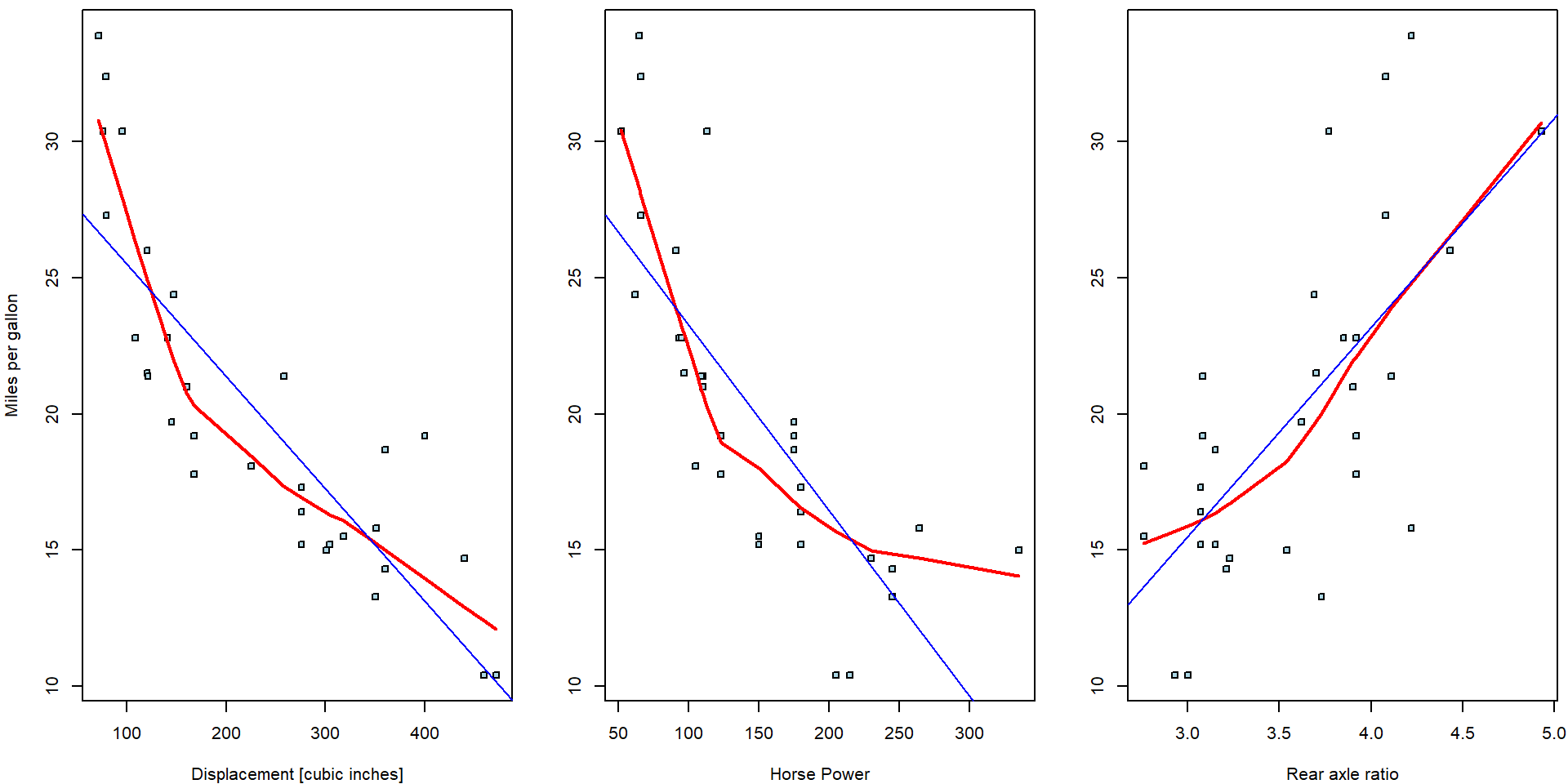
However, the multiple regression model does not estimate a line but a
three dimensional subspace. For instance, the intercept value \(\widehat{\beta}_0 = 29.6\) from the simple
regression model fit_disp stands for the expected driven
miles per one gallon if the displacement of the engine is equal to zero
cubic inches (i.e., an unrealistic scenario).
Nevertheless, the intercept estimate \(\widehat{\beta}_0 = 19.34\) stands for the expected driven miles per one gallon but if all three regressors – the information about the displacement, the horse powers, and, also, the rear to axel ratio – are all equal to zero. Therefore, to draw simple line for two-dimensional scatterplot, it is needed to pre-select some reasonable information (realistic case) for all other covariates in the model.
For instance, consider the multiple model from above and, in particular, the scatterplot of the consumption against the displacement. Various regression lines can be considered:
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(mpg ~ disp, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue",
xlab = "Displacement [cubic inches]", ylab = "Miles per gallon")
xGrid <- seq(min(mtcars$disp), max(mtcars$disp, 100))
mCoeff <- fit1$coeff
yMeanMean <- mCoeff[1] + mCoeff[2] * xGrid + mCoeff[3] * mean(mtcars$hp) + mCoeff[4] * mean(mtcars$drat)
lines(yMeanMean ~ xGrid, col = "red", lwd = 2)
yMinMin <- mCoeff[1] + mCoeff[2] * xGrid + mCoeff[3] * min(mtcars$hp) + mCoeff[4] * min(mtcars$drat)
yMaxMax<- mCoeff[1] + mCoeff[2] * xGrid + mCoeff[3] * max(mtcars$hp) + mCoeff[4] * max(mtcars$drat)
lines(yMinMin ~ xGrid, col = "blue", lwd = 2, lty = 2)
lines(yMaxMax ~ xGrid, col = "darkgreen", lwd = 2, lty = 2)
legend("topright",
legend = c("Average HP & average rear axle ratio",
"Min HP & min rear axle ratio",
"Max HP & max rear axle ratio"),
col =c("red", "blue", "darkgreen"), lwd = c(3,3,3), lty = c(1,2,2))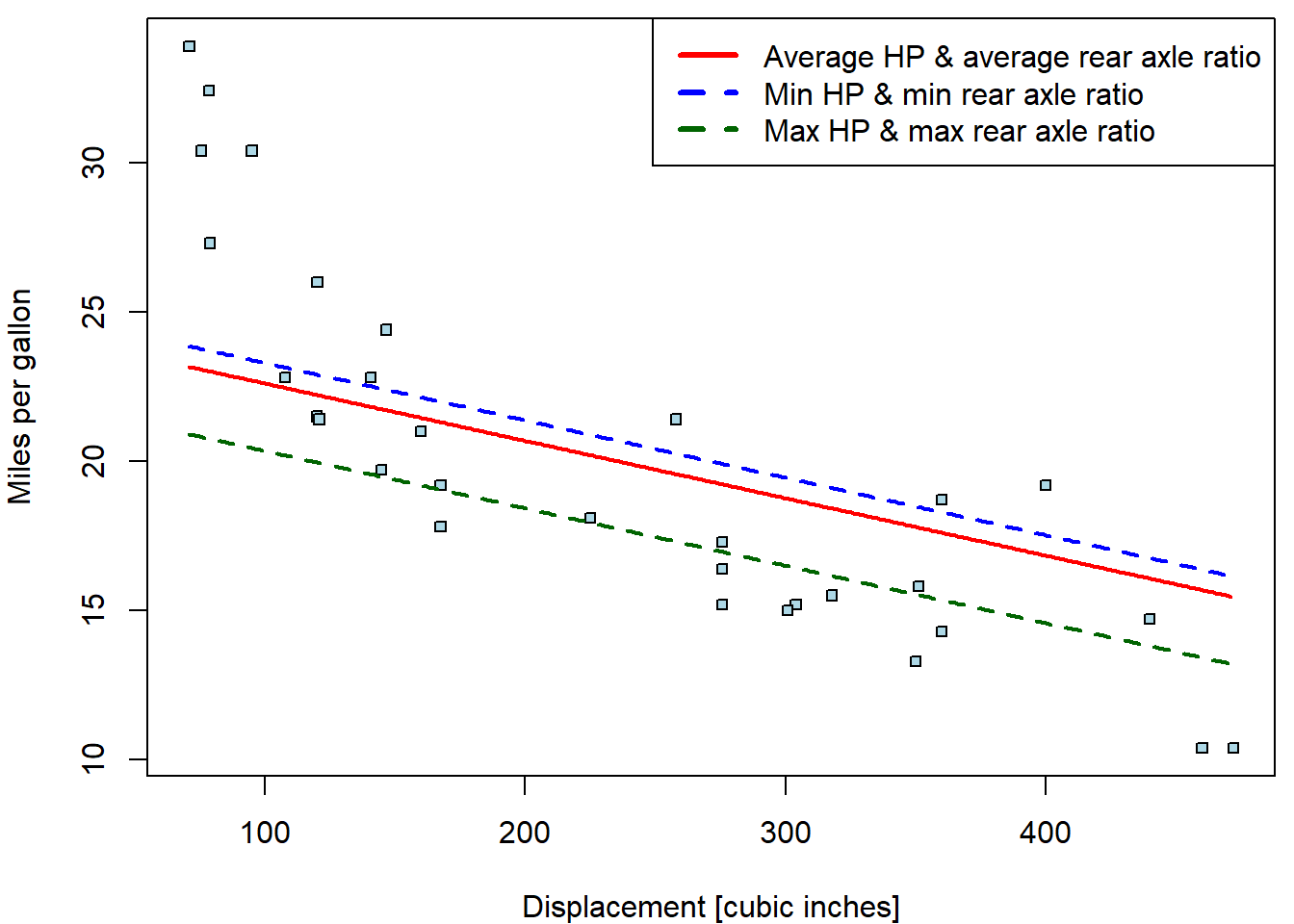
Individual work
- The whole set of different regression lines can be plotted depending on particular choices for the remaining covariates that are not used in the scatterplot (in this case, the information about the horse powers and the rear axle ratio).
- For all three considered covariates draw a separate scatterplot with a set of regression lines that will reasonably explain the underlying dependence of the car consumption.
- In practical applications, it is reasonable to use the exploratory analysis to pre-select some reasonable values for the covariates that are used in the model formula, the relationship of the dependent covariate is not directly illustrated in the plot. For instance, one can use a series of nice curves for the estimated deciles of, let’s say the horse power, while always using the median value of the rear axle ratio.
2. Basic inference on \(\boldsymbol{\beta} \in \mathbb{R}^p\)
The main purpose of statistic is to do inference – i.e., to say something relevant about the whole (unknown) population (from which we have a small random sample \(\{(Y_i, \boldsymbol{X}_i^\top)^\top;~i = 1, \dots, 32\}\)) using only the information (data) in the sample and the techniques of statistical inference. There are two main tools for the statistical inference – confidence intervals for the unknown parameters and the hypothesis tests.
While the confidence intervals must be calculated manually – however,
using just the standard output from the regression model – some
statistical tests are provided automatically. A statistical output of
the multiple linear regression model fitted above can be obtained by the
command summary():
summary(fit1)##
## Call:
## lm(formula = mpg ~ disp + hp + drat, data = mtcars)
##
## Residuals:
## Min 1Q Median 3Q Max
## -5.122 -1.845 -0.446 1.134 6.496
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 19.34429 6.37088 3.04 0.0051 **
## disp -0.01923 0.00937 -2.05 0.0496 *
## hp -0.03123 0.01334 -2.34 0.0266 *
## drat 2.71498 1.48737 1.83 0.0786 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.01 on 28 degrees of freedom
## Multiple R-squared: 0.775, Adjusted R-squared: 0.751
## F-statistic: 32.2 on 3 and 28 DF, p-value: 3.28e-09Four (the number of estimated parameters) statistical tests – \(p\)-values of the tests are provided in the output (the fourth column of the table) by default. In each case, the corresponding null hypothesis states that the estimated parameter (not the estimate, but the unknown true value) is equal to zero, i.e.,
\[ H_0: \beta_j = 0 \] against a general alternative \[ H_A: \beta_j \neq 0 \] where \(j \in \{1, 2, 3, 4\}\) corresponds to the line in the output table, respectively the element of the estimated parameter vector \(\boldsymbol{\beta} \in \mathbb{R}^4\).
Individual work
- Recall, how a standard statistical test is provided. What is the corresponding test statistic, the distribution of the test statistic under the null hypothesis, or the critical region for the test?
- Can you use some of the information provided in the output to reconstruct the quantities needed for the test (e.g., the test statistic?).
- What should be some reasonable assumptions to be considered together with the test?
- What is the interpretation of the statistical test given in the
first line of the summary output (the line denoted as
Intercept)? - What is the interpretation of the statistical tests for the
remaining three lines of the summary output (the lines denoted as
disp,hp, anddrat)?
3. K-variate parametrizations of a numeric covariate
On previous exercise class we considered transformations that changed a single covariate into another single covariate. These are typically:
- shift (by a common value)
- scale (g to kg, km to miles, etc.)
- categorical (two groups)
- square root
- logarithmic transformation (will be covered on exercise 6 in more detail)
- …
But one covariate could be parametrized by more than 1 column! Typical examples:
- quadratic trend
- polynomial trend (will be covered on exercise 7 in more detail)
- piece-wise constant = categorical parametrization (exercise 5)
- continuous piece-wise linear trend
- spline basis (will be covered on exercise 7 in more detail)
Consider again the relationship between consumption and displacement:
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(mpg ~ disp, data = mtcars, pch = 22, cex = 0.8, bg = "lightblue", ylab = "Miles per gallon", xlab = "Displacement [cubic inches]")
lines(lowess(mtcars$mpg ~ mtcars$disp), col = "red", lwd = 2)
abline(fit_disp, col = "blue")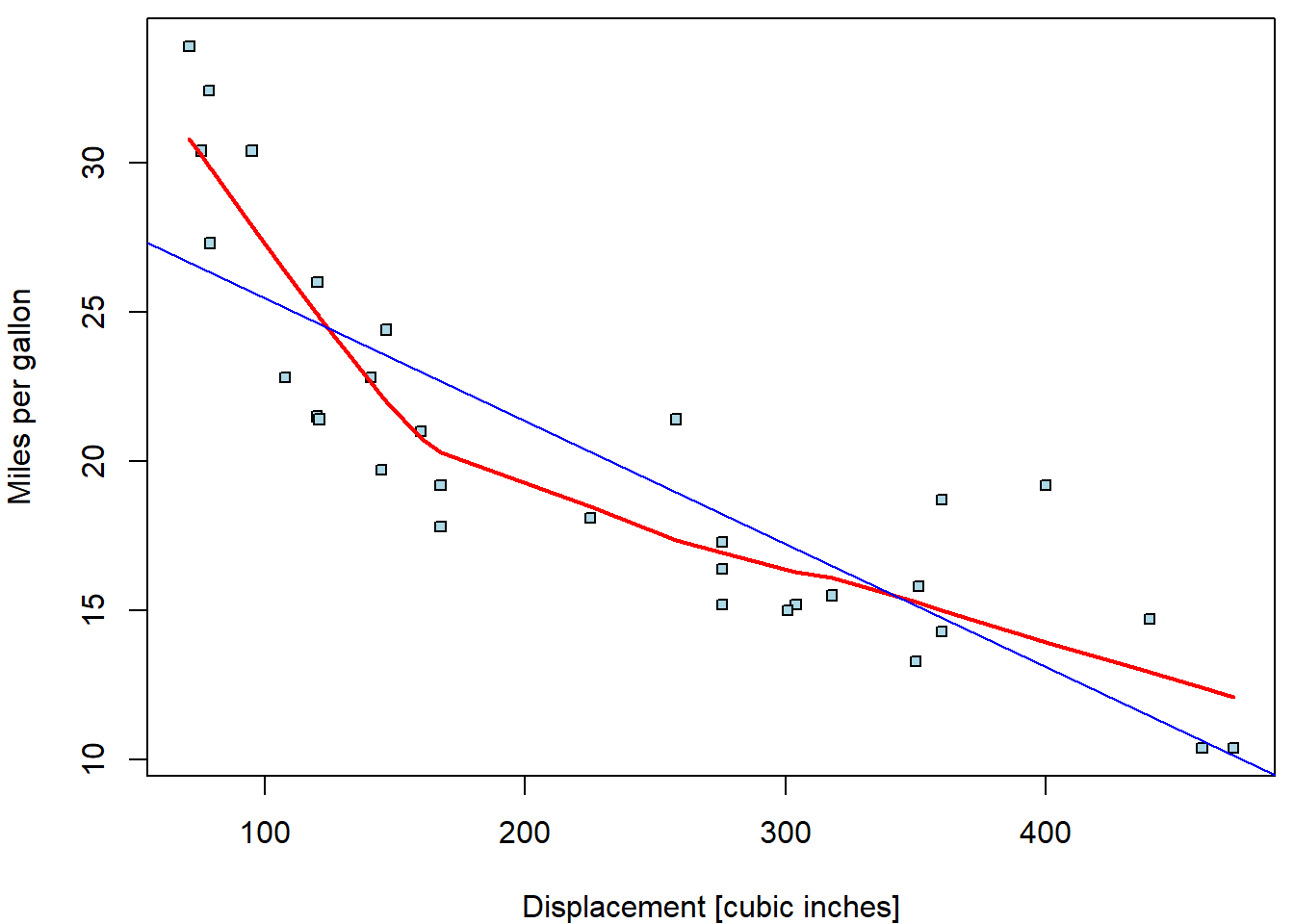
Apparently, linear trend describes the relationship poorly. The
lowess curve suggests more concave behaviour. Let us
consider other possible parametrizations (square root, quadratic
polynomial, cubic polynomial, piecewise-linear function with break in
170):
fit_disp <- list()
fit_disp[["line"]] <- lm(mpg ~ disp, data = mtcars)
fit_disp[["sqrt"]] <- lm(mpg ~ sqrt(disp), data = mtcars)
fit_disp[["log2"]] <- lm(mpg ~ log2(disp), data = mtcars)
fit_disp[["quad"]] <- lm(mpg ~ disp + disp^2, data = mtcars) # wrong!!!
fit_disp[["quad"]] <- lm(mpg ~ disp + I(disp^2), data = mtcars) # this is correct!
fit_disp[["cube"]] <- lm(mpg ~ disp + I(disp^2) + I(disp^3), data = mtcars)
fit_disp[["hoc1"]] <- lm(mpg ~ pmin(disp-170, 0) + pmax(170, disp), data = mtcars)
fit_disp[["hoc2"]] <- lm(mpg ~ disp + pmax(170, disp), data = mtcars)
fit_disp[["flat"]] <- lm(mpg ~ pmin(170, disp), data = mtcars)
sum_disp <- lapply(fit_disp, summary) # summary for each model in the listNotice that the additional power terms of disp have to
be wrapped by I(). The reason is that ^2 has
different meaning in formula notation (interactions).
Functions min and max return always only
one number - minimum or maximum from all given numbers. However, we wish
to compute minimum or maximum in vectorized form, always from two
numbers. This is done with pmin and pmax.
The models fit_disp_hoc1 and fit_disp_hoc2
are two equivalent parametrizations of the same function. In the first
one, beta coefficients (besides intercept) correspond to the slopes in
different intervals. \[
f(x) = \left\{
\begin{aligned}
a_1 + b_1 x, & \quad \text{for}\; x \in (-\infty, 170], \\
a_2 + b_2 x, & \quad \text{for}\; x \in [170, \infty).
\end{aligned}
\right.
\quad \Longrightarrow \quad
f(x) = a_2 + b_1 \min (x-170, 0) + b_2 \max(170, x)
\] The second parametrization, the second coefficient is the
slope in the first interval \((-\infty,
170]\) and the third coefficient describes the change from this
slope to the slope in the other interval \([170, \infty)\). \[
f(x) = \left\{
\begin{aligned}
a_1 + b x, & \quad \text{for}\; x \in (-\infty, 170], \\
a_2 + (b + \delta) x, & \quad \text{for}\; x \in [170, \infty).
\end{aligned}
\right.
\quad \Longrightarrow \quad
f(x) = a_2 + b x + \delta \max(170, x)
\] The intercept is the same in both cases as it corresponds to
the intercept on the second interval. Compare:
sum_disp[["hoc1"]]##
## Call:
## lm(formula = mpg ~ pmin(disp - 170, 0) + pmax(170, disp), data = mtcars)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.595 -1.680 0.067 1.115 4.866
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 22.17664 1.66970 13.28 7.4e-14 ***
## pmin(disp - 170, 0) -0.12180 0.01569 -7.77 1.5e-08 ***
## pmax(170, disp) -0.01961 0.00535 -3.67 0.00098 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.36 on 29 degrees of freedom
## Multiple R-squared: 0.856, Adjusted R-squared: 0.846
## F-statistic: 86.2 on 2 and 29 DF, p-value: 6.26e-13sum_disp[["hoc2"]]##
## Call:
## lm(formula = mpg ~ disp + pmax(170, disp), data = mtcars)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.595 -1.680 0.067 1.115 4.866
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 22.1766 1.6697 13.28 7.4e-14 ***
## disp -0.1218 0.0157 -7.77 1.5e-08 ***
## pmax(170, disp) 0.1022 0.0194 5.26 1.2e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.36 on 29 degrees of freedom
## Multiple R-squared: 0.856, Adjusted R-squared: 0.846
## F-statistic: 86.2 on 2 and 29 DF, p-value: 6.26e-13See? Intercept rows are both \(22.17664\). Next, \(-0.1218 + 0.10219 = -0.019607\)
The last model fit_disp_flat has some slope in the first
interval, but after 170 there is no trend only constant.
In order to plot the fitted curves we define the functions:
fun <- list()
fun[["line"]] <- function(beta, x){beta[1] + beta[2] * x}
fun[["sqrt"]] <- function(beta, x){beta[1] + beta[2] * sqrt(x)}
fun[["log2"]] <- function(beta, x){beta[1] + beta[2] * log2(x)}
fun[["quad"]] <- function(beta, x){beta[1] + beta[2] * x + beta[3] * x^2}
fun[["cube"]] <- function(beta, x){beta[1] + beta[2] * x + beta[3] * x^2 + beta[4] * x^3}
fun[["hoc1"]] <- function(beta, x){beta[1] + beta[2] * pmin(x-170, 0) + beta[3] * pmax(170, x)}
fun[["hoc2"]] <- function(beta, x){beta[1] + beta[2] * x + beta[3] * pmax(170, x)}
fun[["flat"]] <- function(beta, x){beta[1] + beta[2] * pmin(170, x)}Now we plot all the fitted curves into one plot:
xgrid <- seq(min(mtcars$disp), max(mtcars$disp), length.out = 201)
COL <- c("blue", "purple", "orchid", "green", "darkgreen", "orange", "red", "peru")
names(COL) <- names(fit_disp)
nice_labels <- c("line", "square root", expression(log[2]),
"quadratic", "cubic",
"hockey 1", "hockey 2", "flat hockey")
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(mpg ~ disp, data = mtcars,
xlab = "Displacement [cubic inches]", ylab = "Miles per gallon",
pch = 22, cex = 0.8, bg = "lightblue")
abline(v = 170, col = "grey", lty = 3)
# now add the estimated curves
for(curve in names(fit_disp)){
lines(xgrid, fun[[curve]](coef(fit_disp[[curve]]), xgrid),
col = COL[curve], lwd = 2, lty = ifelse(curve == "hoc2", 2, 1))
}
legend("topright", legend = nice_labels, bty = "n",
col = COL, lty = c(1,1,1,1,1,2,1), lwd = 2)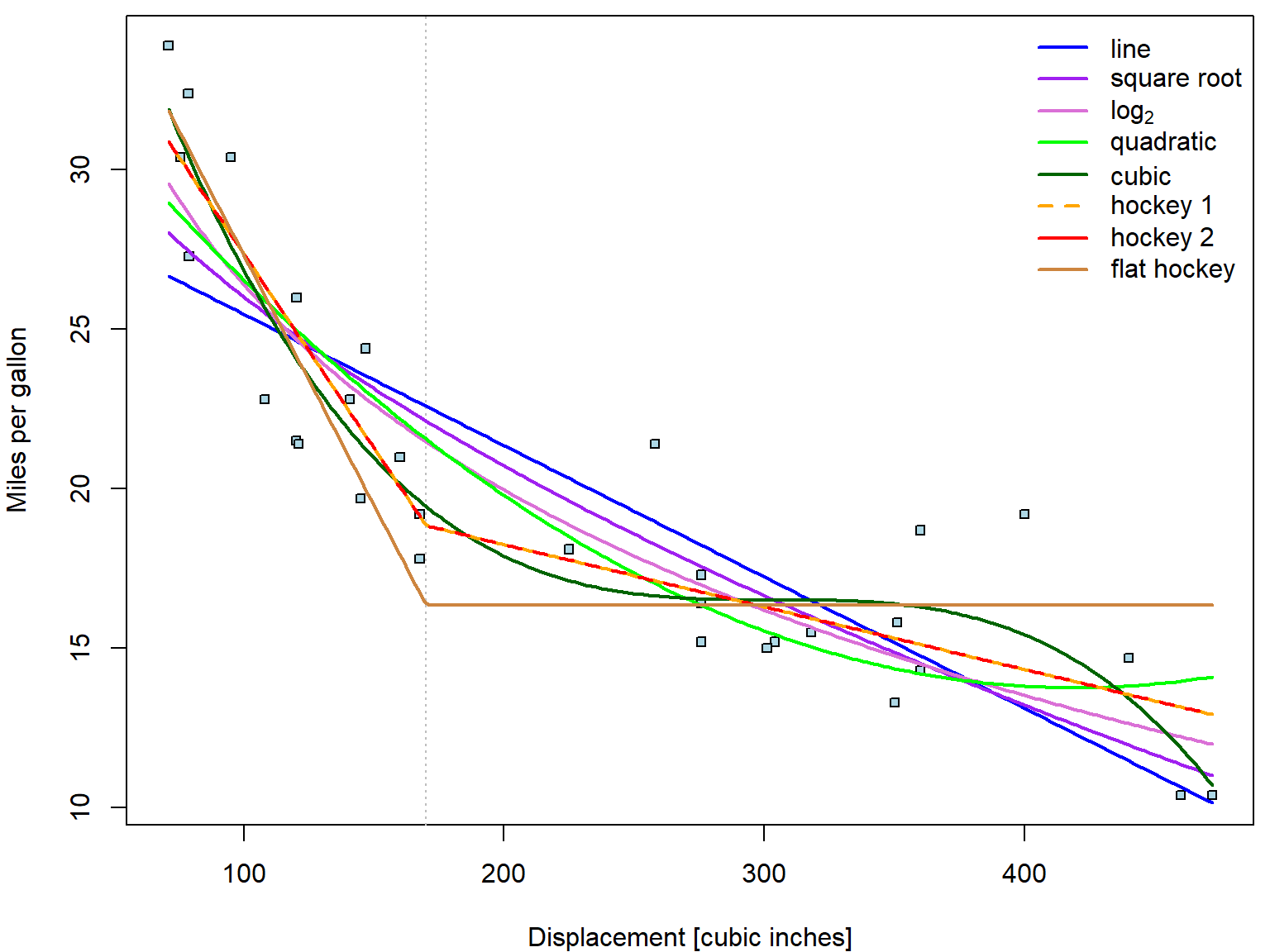
You actually do not have to define the functions on your own. There
is always an option to use predict function to estimate the
fitted value fit given grid of newdata
covariates.
pred_disp <- lapply(fit_disp, predict, newdata = data.frame(disp = xgrid))The predict function will work this way as long as disp
is transformed directly within the model formula. If you would define
disp2 = disp^2, then your newdata need to
contain disp2 as well. See? The results are the same:
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(mpg ~ disp, data = mtcars,
xlab = "Displacement [cubic inches]", ylab = "Miles per gallon",
pch = 22, cex = 0.8, bg = "lightblue")
abline(v = 170, col = "grey", lty = 3)
# now add the estimated curves
for(curve in names(fit_disp)){
lines(xgrid, pred_disp[[curve]],
col = COL[curve], lwd = 2, lty = ifelse(curve == "hoc2", 2, 1))
}
legend("topright", legend = nice_labels, bty = "n",
col = COL, lty = c(1,1,1,1,1,2,1), lwd = 2)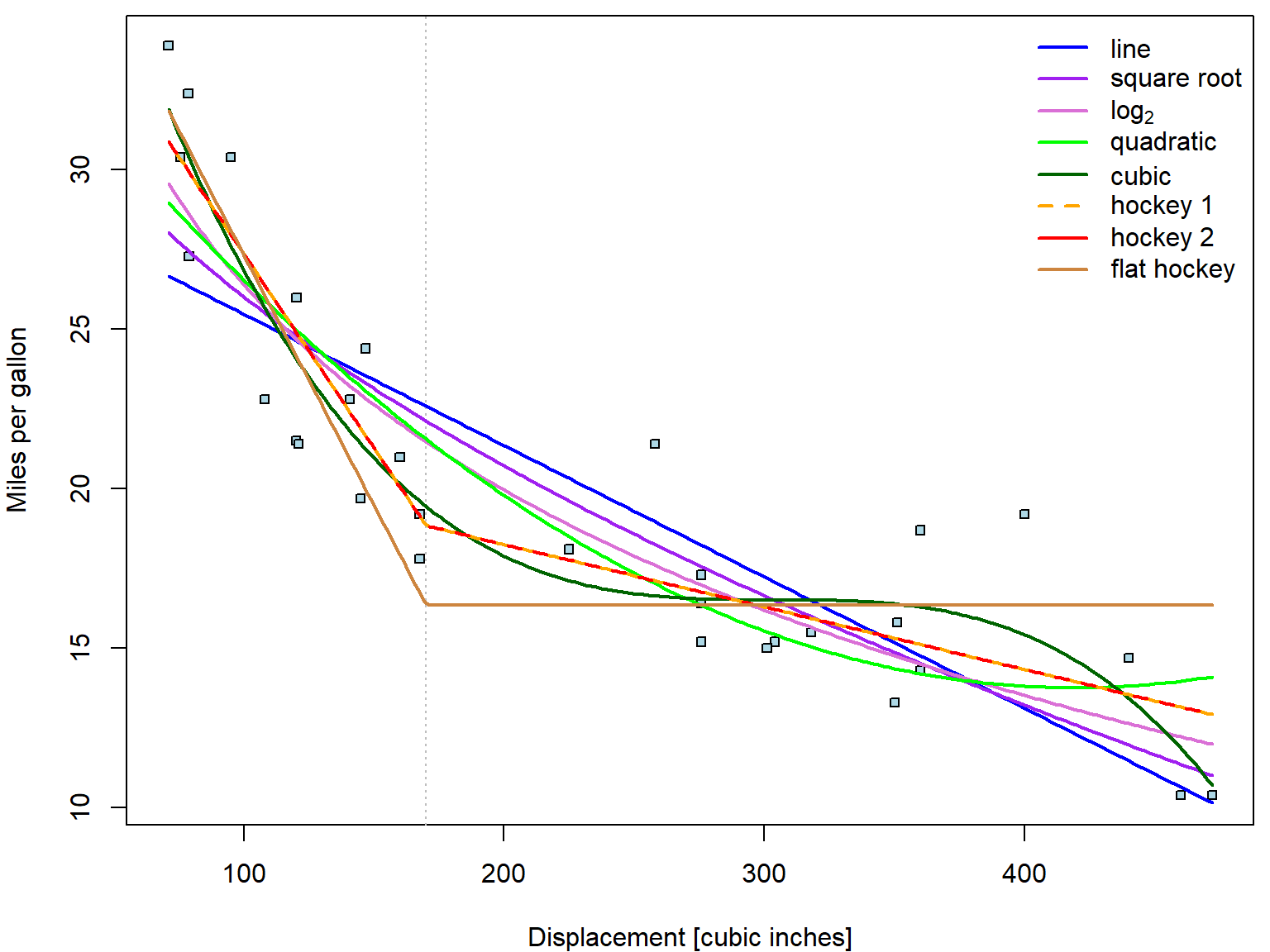
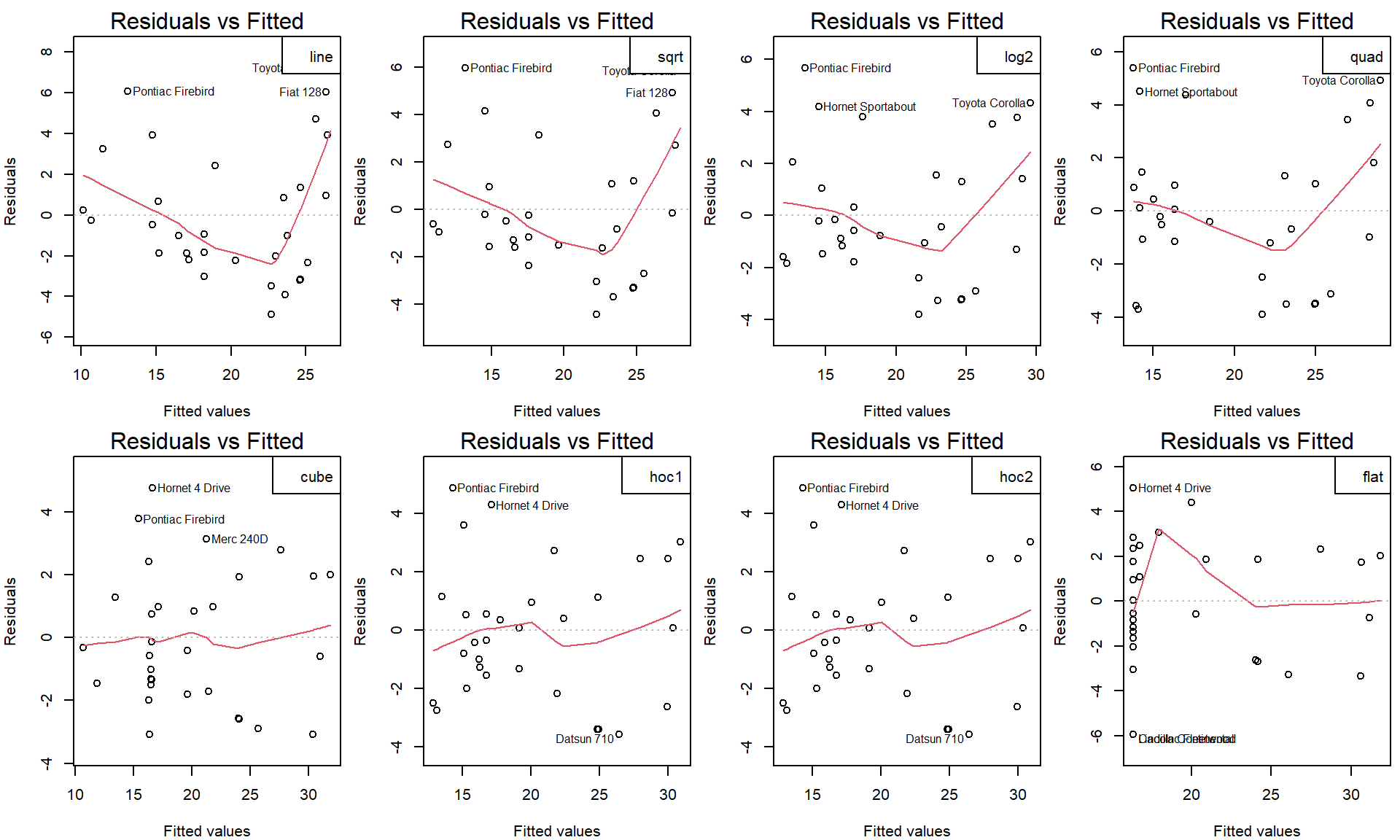
How do we choose suitable transformation?
- Visual appeal (above),
- Satisfactory visual diagnostic tools (will be covered in more details later in exercise 7)
- Proportion of explained variability - by \(R^2\) (always higher for model compared to its submodel),
- Overall F-test p-value (takes number of coefficients required into account),
- Model selection criteria (AIC, BIC) - to avoid over-fitting,
- Submodel tests (if valid)
table <- data.frame(R2 = unlist(lapply(sum_disp, function(s){s$r.squared})),
adjR2 = unlist(lapply(sum_disp, function(s){s$adj.r.squared})),
Fpval = unlist(lapply(sum_disp, function(s){
pf(s$fstatistic["value"], df1 = s$fstatistic["numdf"],
df2 = s$fstatistic["dendf"], lower.tail = FALSE
)})),
AIC = unlist(lapply(fit_disp, AIC)))
library(knitr)
kable(table, digits = 15)| R2 | adjR2 | Fpval | AIC | |
|---|---|---|---|---|
| line | 0.7183 | 0.7090 | 9.380e-10 | 170.2 |
| sqrt | 0.7756 | 0.7681 | 2.993e-11 | 162.9 |
| log2 | 0.8229 | 0.8169 | 8.400e-13 | 155.4 |
| quad | 0.7927 | 0.7784 | 1.229e-10 | 162.4 |
| cube | 0.8771 | 0.8639 | 7.350e-13 | 147.7 |
| hoc1 | 0.8560 | 0.8461 | 6.260e-13 | 150.7 |
| hoc2 | 0.8560 | 0.8461 | 6.260e-13 | 150.7 |
| flat | 0.7892 | 0.7822 | 1.162e-11 | 160.9 |
par(mfrow = c(2,4), mar = c(4,4,2,0.5))
for(curve in names(fit_disp)){
plot(fit_disp[[curve]], which = 1)
legend("topright", legend = curve)
}
Individual work
- Choose the best fitting trend.
- Repeat for other numeric covariates -
hpanddrat. - Fit a model with the chosen parametrizations of all three covariates.