Introduction to fitting linear models
Jan Vávra
Exercise 2
Download this R markdown as: R, Rmd.
Loading the data and libraries
library("mffSM")
library("colorspace")We want to examine a nitrogen concentration measured at various
depths of the peat. Load the data peat below and try to
understand the design of the underlying experiment. Focus on various
depths in particular.
peat <- read.csv("http://www.karlin.mff.cuni.cz/~vavraj/nmfm334/data/peat.csv",
header = TRUE, stringsAsFactors = TRUE)dim(peat)## [1] 148 3str(peat)## 'data.frame': 148 obs. of 3 variables:
## $ group: Factor w/ 4 levels "CB","CB-VJJ",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ N : num 0.738 0.847 1.063 1.142 1.272 ...
## $ depth: int 2 4 6 8 10 12 14 2 4 6 ...head(peat)## group N depth
## 1 VJJ 0.7384 2
## 2 VJJ 0.8474 4
## 3 VJJ 1.0630 6
## 4 VJJ 1.1419 8
## 5 VJJ 1.2722 10
## 6 VJJ 1.4033 12summary(peat)## group N depth
## CB :35 Min. :0.499 Min. : 0.00
## CB-VJJ:40 1st Qu.:0.899 1st Qu.: 4.00
## VJJ :33 Median :1.034 Median : 8.00
## VJJ-CB:40 Mean :1.077 Mean : 7.53
## 3rd Qu.:1.244 3rd Qu.:12.00
## Max. :1.692 Max. :14.00Visual exploration
Basic scatterplot for Nitrogen concentration given depth:
XLAB <- "Depth [cm]"
YLAB <- "Nitrogen concentration [weight %]"
XLIM <- range(peat[, "depth"])
YLIM <- range(peat[, "N"])
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(N ~ depth, data = peat, pch = 21, bg = "lightblue",
xlab = XLAB, ylab = YLAB, xlim = XLIM, ylim = YLIM)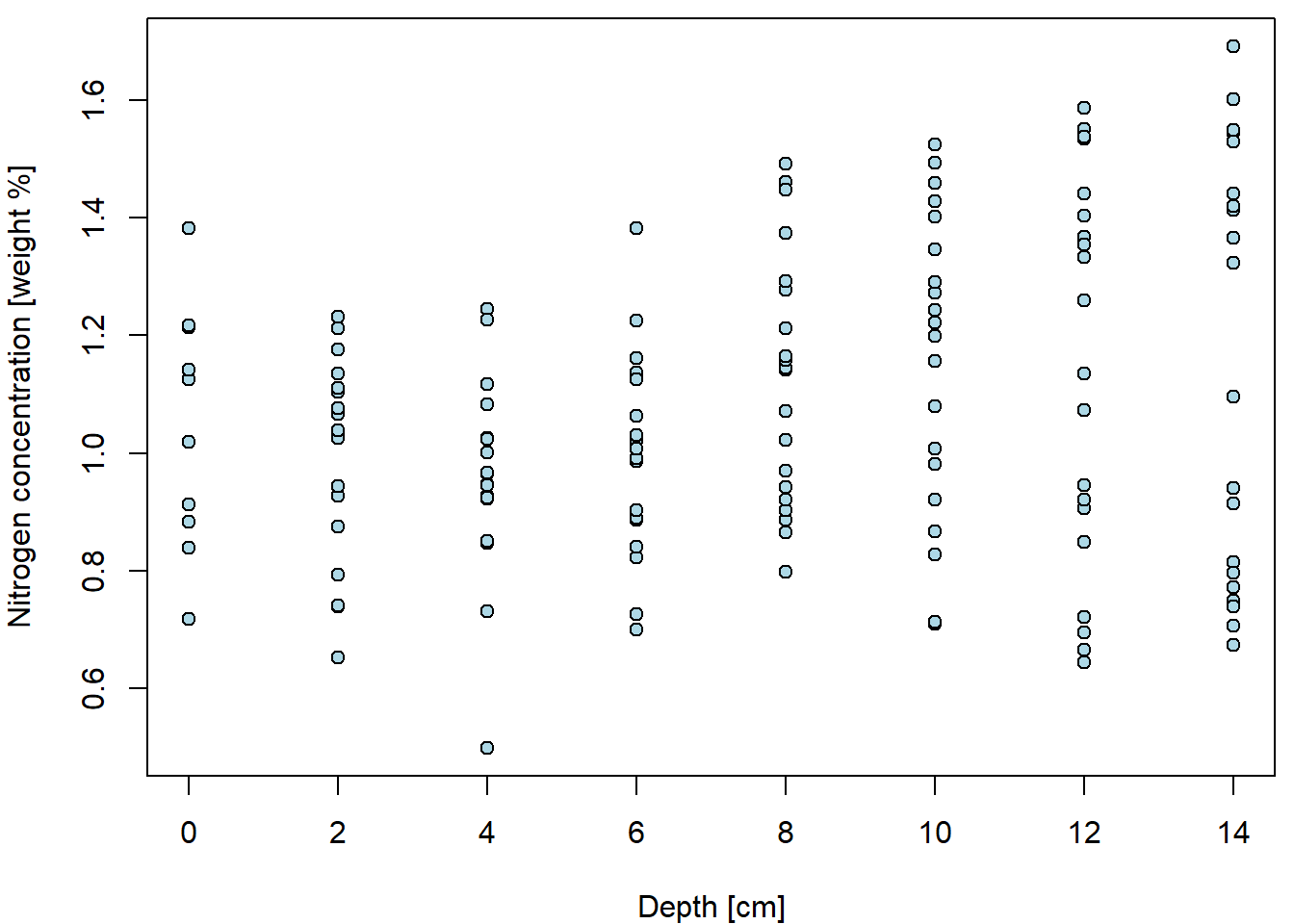
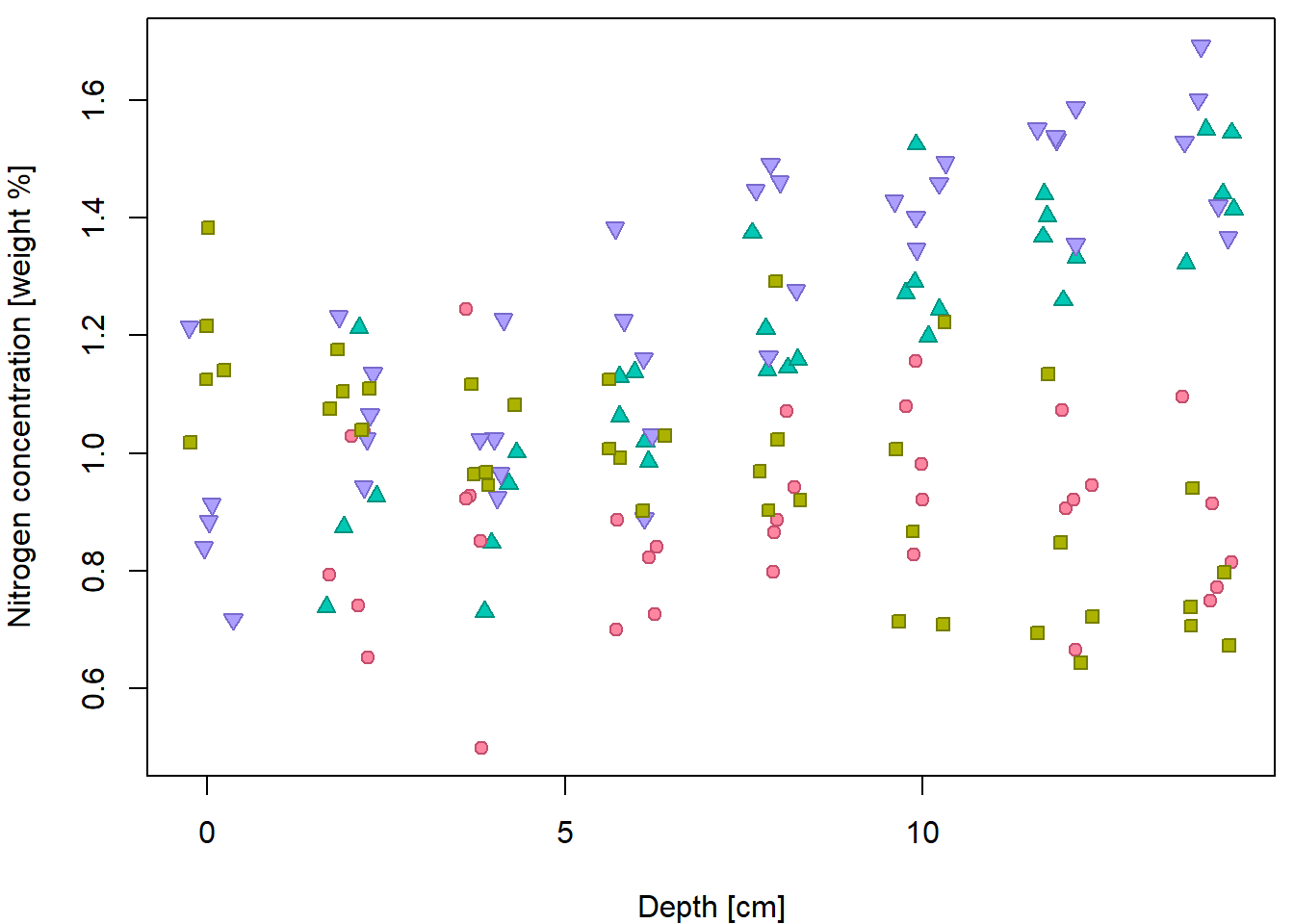
But it can be done better… by distinguishing for different peat types
(variable group).
Groups <- levels(peat[, "group"])
print(Groups)## [1] "CB" "CB-VJJ" "VJJ" "VJJ-CB"PCH <- c(21, 22, 24, 25)
DARK <- rainbow_hcl(4, c = 80, l = 35)
COL <- rainbow_hcl(4, c = 80, l = 50)
BGC <- rainbow_hcl(4, c = 80, l = 70)
names(PCH) <- names(COL) <- names(BGC) <- Groups
par(mfrow = c(2,2), mar = c(4,4,2,1))
for(g in 1:4){
Group <- Groups[g]
xx <- subset(peat, group == Group)[, "depth"]
yy <- subset(peat, group == Group)[, "N"]
plot(xx, yy, pch = PCH[g], col = COL[g], bg = BGC[g],
xlab = XLAB, ylab = YLAB, main = Group, xlim = XLIM, ylim = YLIM)
} 
Conditional empirical characteristics by groups
tabs <- list()
# g <- 1 # run for one g
for(g in 1:length(Groups)){
# data subset containing only the g-th group
subdata <- subset(peat, group == Groups[g])
# averages for particular depths
Mean <- with(subdata, tapply(N, depth, mean))
# standard deviations for particular depths
StdDev <- with(subdata, tapply(N, depth, sd))
# numbers of observations for particular depths
ns <- with(subdata, tapply(N, depth, length))
# cat("\n",Groups[g],"\n")
# print(Mean)
# print(StdDev)
# print(ns)
tabs[[g]] <- data.frame(Depth = as.numeric(names(Mean)), Mean = Mean,
Std.Dev = StdDev, n = ns)
rownames(tabs[[g]]) <- 1:nrow(tabs[[g]])
}
names(tabs) <- Groups
print(tabs)## $CB
## Depth Mean Std.Dev n
## 1 2 0.8504 0.17437 5
## 2 4 0.8886 0.26575 5
## 3 6 0.7951 0.07906 5
## 4 8 0.9123 0.10257 5
## 5 10 0.9931 0.12951 5
## 6 12 0.9025 0.14771 5
## 7 14 0.8694 0.14203 5
##
## $`CB-VJJ`
## Depth Mean Std.Dev n
## 1 0 1.1769 0.13512 5
## 2 2 1.1012 0.05042 5
## 3 4 1.0153 0.07849 5
## 4 6 1.0114 0.08000 5
## 5 8 1.0216 0.15842 5
## 6 10 0.9036 0.21678 5
## 7 12 0.8086 0.19743 5
## 8 14 0.7710 0.10511 5
##
## $VJJ
## Depth Mean Std.Dev n
## 1 2 0.9381 0.19973 4
## 2 4 0.8817 0.11941 4
## 3 6 1.0673 0.06671 5
## 4 8 1.2065 0.09810 5
## 5 10 1.3062 0.12734 5
## 6 12 1.3611 0.06927 5
## 7 14 1.4550 0.09526 5
##
## $`VJJ-CB`
## Depth Mean Std.Dev n
## 1 0 0.9132 0.18372 5
## 2 2 1.0804 0.10998 5
## 3 4 1.0331 0.11638 5
## 4 6 1.1380 0.18807 5
## 5 8 1.3686 0.14144 5
## 6 10 1.4260 0.05604 5
## 7 12 1.5132 0.09093 5
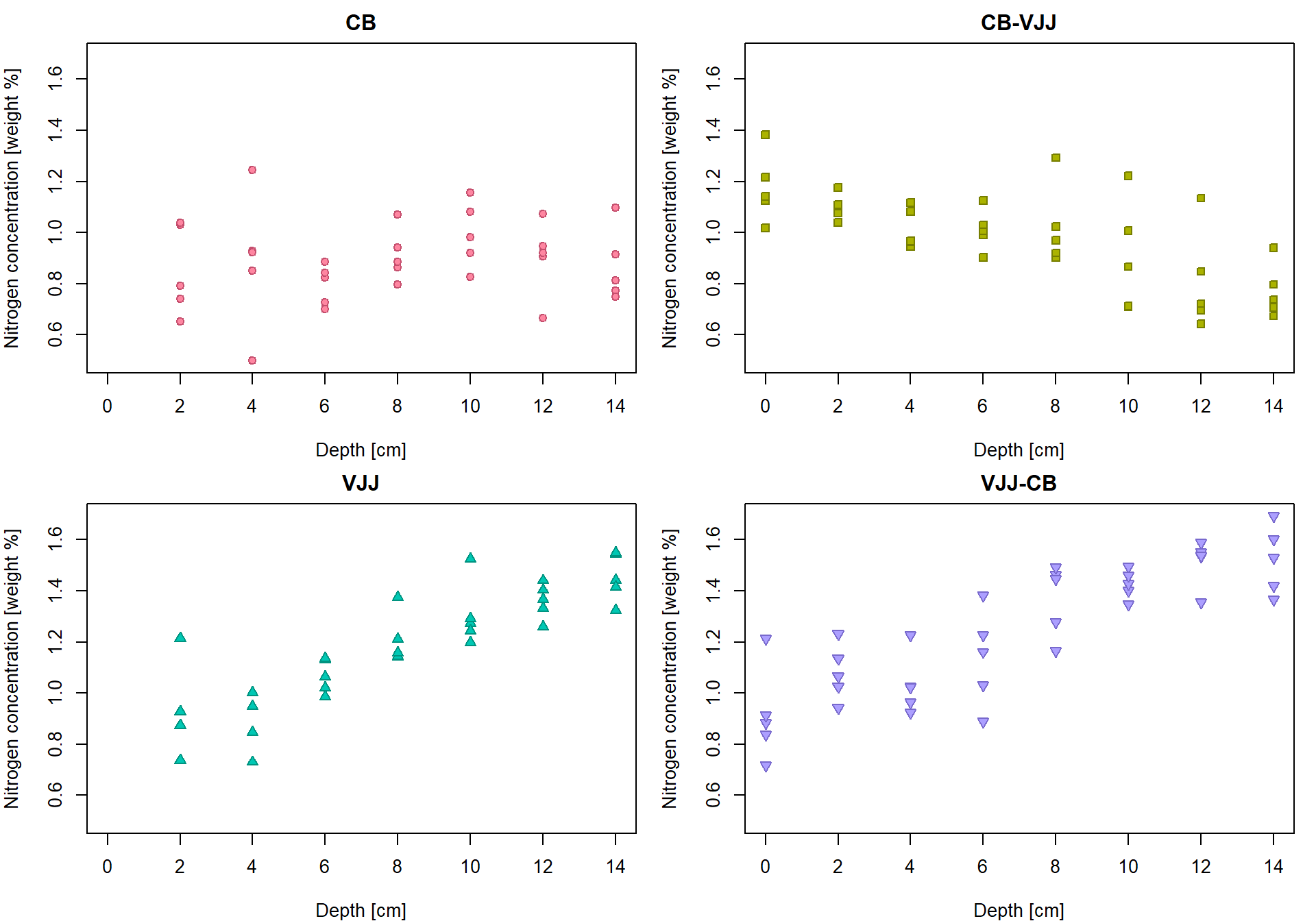
## 8 14 1.5221 0.13198 5Insert the group averages into the original plot
par(mfrow = c(2,2), mar = c(4,4,2,1))
for (g in 1:4){
Group <- Groups[g]
xx <- subset(peat, group == Group)[, "depth"]
yy <- subset(peat, group == Group)[, "N"]
plot(xx, yy, pch = PCH[g], col = COL[g], bg = BGC[g],
xlab = XLAB, ylab = YLAB, main = Group, xlim = XLIM, ylim = YLIM)
lines(tabs[[g]][, "Depth"], tabs[[g]][, "Mean"], type = "b",
pch = 23, lty = 2, cex = 2, col = COL[g], bg = BGC[g])
} 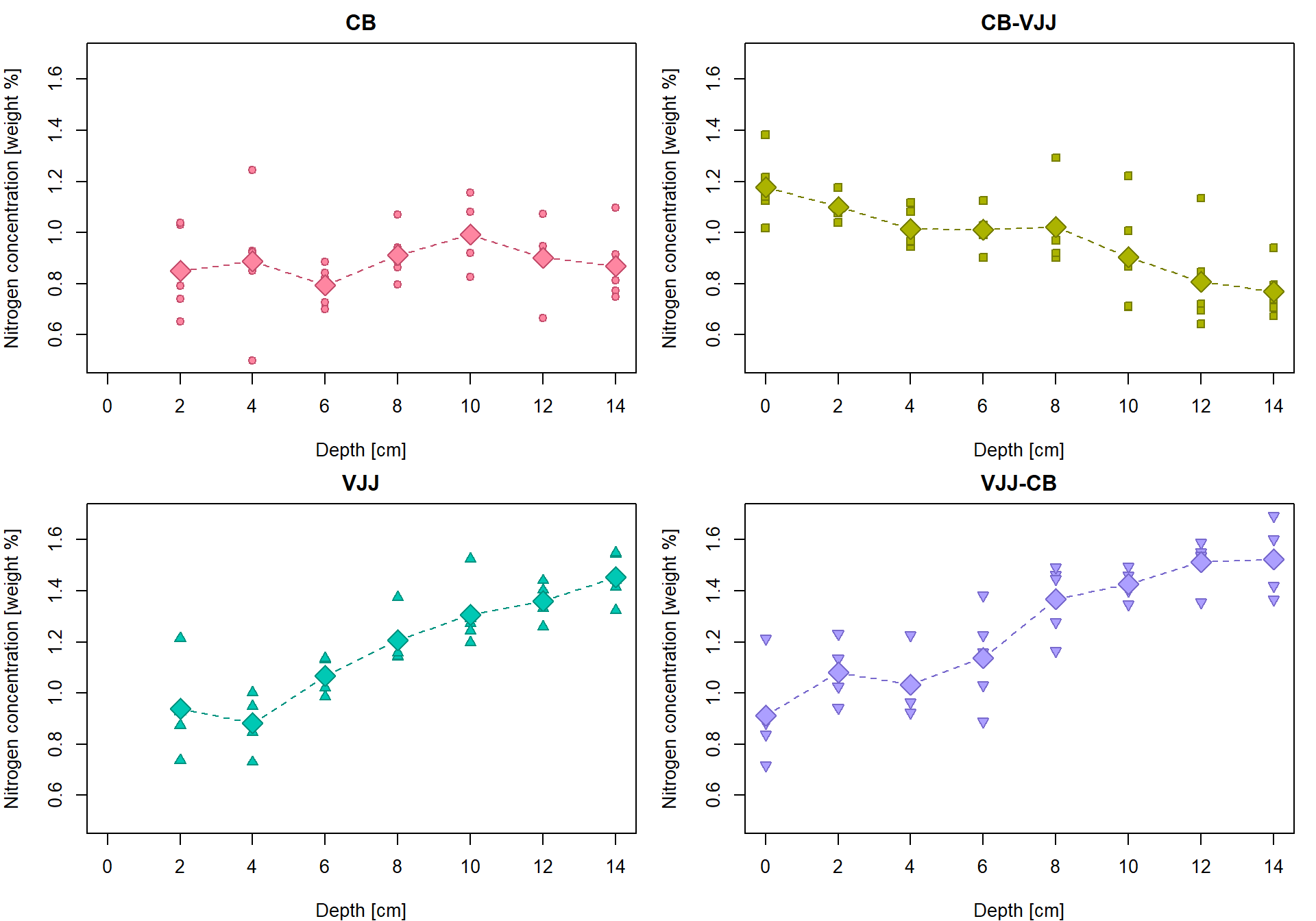
Linear regression line for the group “CB-VJJ”
Let’s fit the least-squares estimated of the regression line
parameters with lm:
Group <- "CB-VJJ"
g <- 2
fit1 <- lm(N ~ depth, data = peat, subset = (group == Group))
print(fit1)##
## Call:
## lm(formula = N ~ depth, data = peat, subset = (group == Group))
##
## Coefficients:
## (Intercept) depth
## 1.1691 -0.0276The regression line establishes a simple relationship between the nitrogen concentration (Y values) and the depth of the measurement (X values).
Yvalues <- peat$N[peat$group == Group]
Xvalues <- peat$depth[peat$group == Group]
Y <- function(a, b, x){
# a simple line parametrization with the intercept 'a' and the slope `b`
return(a + b * x)
}
par(mfrow = c(1,1), mar = c(4,4,2,1))
# the underlying data
plot(Yvalues ~ Xvalues, pch = PCH[Group], col = COL[Group], bg = BGC[Group],
xlab = XLAB, ylab = YLAB, main = Group, xlim = XLIM, ylim = YLIM)
# empirical group means
lines(tabs[[g]][, "Depth"], tabs[[g]][, "Mean"], type = "b",
pch = 23, lty = 2, cex = 2, col = COL[g], bg = BGC[g])
# regression line given by the LS estimates (for the specific values of the depth)
points(Y(fit1$coeff[1], fit1$coeff[2], Xvalues) ~ Xvalues,
pch = 16, col = DARK[g], bg = BGC[g], cex = 1.5)
# regression line by the 'lm()' function (for the whole real line)
abline(fit1, col = DARK[g], lwd = 2)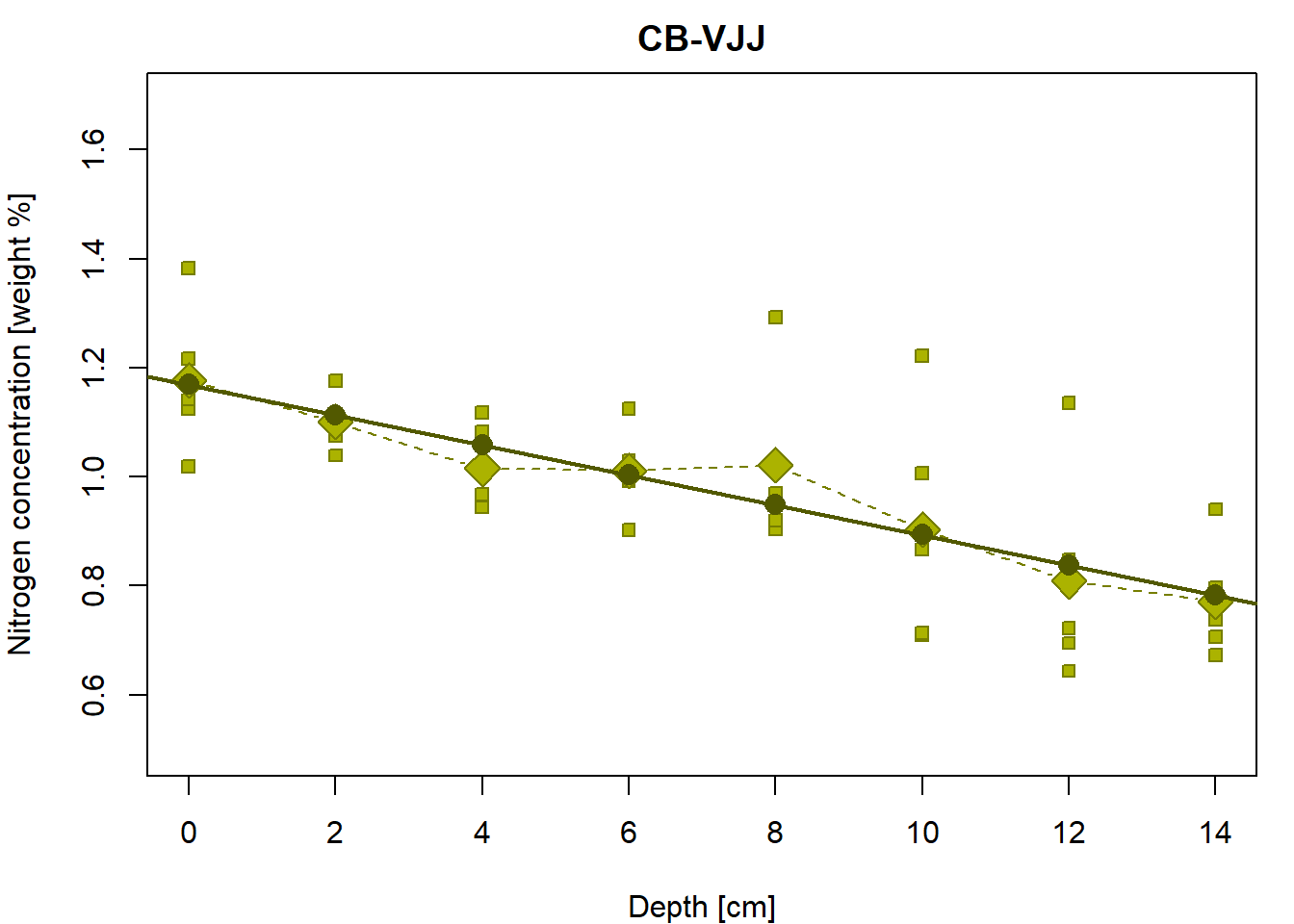 Object
Object fit1 contains many interesting quantities…
fit1 is in fact a list and its components are accessible
either as fit1[["COMPONENT"]] or as
fit1$COMPONENT
names(fit1)## [1] "coefficients" "residuals" "effects" "rank" "fitted.values" "assign"
## [7] "qr" "df.residual" "xlevels" "call" "terms" "model"Estimated coefficients (intercept and slope):
fit1[["coefficients"]]## (Intercept) depth
## 1.16907 -0.02755fit1$coefficients## (Intercept) depth
## 1.16907 -0.02755coef(fit1) # hat{beta}## (Intercept) depth
## 1.16907 -0.02755Fitted values:
fit1[["fitted.values"]]## 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123
## 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384
## 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138
## 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935
## 139 140 141 142 143 144 145 146 147 148
## 0.8384 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384 0.7833fitted(fit1) # hat{Y}## 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123
## 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384
## 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138
## 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935
## 139 140 141 142 143 144 145 146 147 148
## 0.8384 0.7833 1.1691 1.1140 1.0589 1.0038 0.9487 0.8935 0.8384 0.7833Yhat = cbind(1,Xvalues) %*% coef(fit1)
all.equal(as.numeric(Yhat), unname(fitted(fit1))) ## [1] TRUEResiduals:
fit1[["residuals"]]## 109 110 111 112 113 114 115 116 117 118 119
## -0.043626 -0.009146 -0.113661 -0.101298 -0.045701 -0.184711 -0.194907 -0.110234 -0.150839 -0.003993 0.023492
## 120 121 122 123 124 125 126 127 128 129 130
## -0.012196 0.074187 0.113269 0.009912 0.013481 0.047742 -0.074664 -0.094068 0.121601 0.343741 0.329025
## 131 132 133 134 135 136 137 138 139 140 141
## 0.296565 0.157088 0.213630 0.062043 -0.091651 0.003871 0.020786 -0.026919 -0.143776 -0.076928 -0.027662
## 142 143 144 145 146 147 148
## -0.038311 0.058281 0.026092 -0.028355 -0.180401 -0.116814 -0.044944residuals(fit1) # U = Y - hat{Y}## 109 110 111 112 113 114 115 116 117 118 119
## -0.043626 -0.009146 -0.113661 -0.101298 -0.045701 -0.184711 -0.194907 -0.110234 -0.150839 -0.003993 0.023492
## 120 121 122 123 124 125 126 127 128 129 130
## -0.012196 0.074187 0.113269 0.009912 0.013481 0.047742 -0.074664 -0.094068 0.121601 0.343741 0.329025
## 131 132 133 134 135 136 137 138 139 140 141
## 0.296565 0.157088 0.213630 0.062043 -0.091651 0.003871 0.020786 -0.026919 -0.143776 -0.076928 -0.027662
## 142 143 144 145 146 147 148
## -0.038311 0.058281 0.026092 -0.028355 -0.180401 -0.116814 -0.044944(Res = c(Yvalues - Yhat))## [1] -0.043626 -0.009146 -0.113661 -0.101298 -0.045701 -0.184711 -0.194907 -0.110234 -0.150839 -0.003993
## [11] 0.023492 -0.012196 0.074187 0.113269 0.009912 0.013481 0.047742 -0.074664 -0.094068 0.121601
## [21] 0.343741 0.329025 0.296565 0.157088 0.213630 0.062043 -0.091651 0.003871 0.020786 -0.026919
## [31] -0.143776 -0.076928 -0.027662 -0.038311 0.058281 0.026092 -0.028355 -0.180401 -0.116814 -0.044944all.equal(as.numeric(Res),unname(residuals(fit1))) ## [1] TRUERank and degrees of freedom:
fit1[["rank"]] # r## [1] 2fit1[["df.residual"]] # n - r## [1] 38(r = qr(cbind(1,Xvalues))$rank)## [1] 2n = length(Yvalues)
n - r## [1] 38Basic inferential quantities can be extracted from the object
returned by summary of lm fit:
summary(fit1) ##
## Call:
## lm(formula = N ~ depth, data = peat, subset = (group == Group))
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.1949 -0.0923 -0.0196 0.0504 0.3437
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.16907 0.03819 30.61 < 2e-16 ***
## depth -0.02755 0.00456 -6.04 5.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.132 on 38 degrees of freedom
## Multiple R-squared: 0.489, Adjusted R-squared: 0.476
## F-statistic: 36.4 on 1 and 38 DF, p-value: 5.08e-07First summary is simple summary of the residuals:
summary(Res)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -0.1949 -0.0923 -0.0196 0.0000 0.0504 0.3437Residual sum of squares and residual standard error
(SSe <- deviance(fit1)) # SS_e## [1] 0.6651sum((fit1$residuals)^2) # the same## [1] 0.6651sqrt(SSe / (n-r)) # residual standard error## [1] 0.1323Matrix \(\mathsf{MS}_e \cdot (\mathbb{X}^\top \mathbb{X})^{-1}\)
vcov(fit1)## (Intercept) depth
## (Intercept) 0.0014586 -1.459e-04
## depth -0.0001459 2.084e-05Standard error terms (square root of the diagonal)
sqrt(diag(vcov(fit1)))## (Intercept) depth
## 0.038191 0.004565Correlation matrix derived from vcov(fit1)
cov2cor(vcov(fit1))## (Intercept) depth
## (Intercept) 1.0000 -0.8367
## depth -0.8367 1.0000Confidence intervals for regression coefficients
confint(fit1, level= 0.95)## 2.5 % 97.5 %
## (Intercept) 1.09176 1.24638
## depth -0.03679 -0.01831Intercept only model for the group “CB-VJJ”
What is it all good for? Another regression line is used to fit the
same data. What is the main difference between fit1 and
fit0?
fit0 <- lm(N ~ 1, data = peat, subset = (group == Group))
summary(fit0)##
## Call:
## lm(formula = N ~ 1, data = peat, subset = (group == Group))
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.333 -0.114 0.023 0.136 0.406
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.9762 0.0289 33.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.183 on 39 degrees of freedomThe model fits the regression line which is always parallel with the x-axis (thus, the slope is always zero)
Y <- function(a, x){
# a simple line parametrization with the intercept 'a' and zero slope
return(a + 0 * x)
}
# the same Y values for each X value... why?
Y(fit0$coeff[1], Xvalues)## [1] 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762
## [16] 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762
## [31] 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762 0.9762par(mfrow = c(1,1), mar = c(4,4,2,1))
# the underlying data
plot(Yvalues~Xvalues, pch = PCH[Group], col = COL[Group], bg = BGC[Group],
xlab = XLAB, ylab = YLAB, main = Group, xlim = XLIM, ylim = YLIM)
# empirical group means
lines(tabs[[g]][, "Depth"], tabs[[g]][, "Mean"], type = "b",
pch = 23, lty = 2, cex = 2, col = COL[g], bg = BGC[g])
# regression line given by the LS estimates (for the specific values of depth)
points(Y(fit0$coeff[1], Xvalues)~Xvalues,
pch = 16, col = DARK[g], bg = BGC[2], cex = 1.5)
# regression line by the 'lm()' function (for the whole real line)
abline(fit0, col = DARK[g], lwd = 2)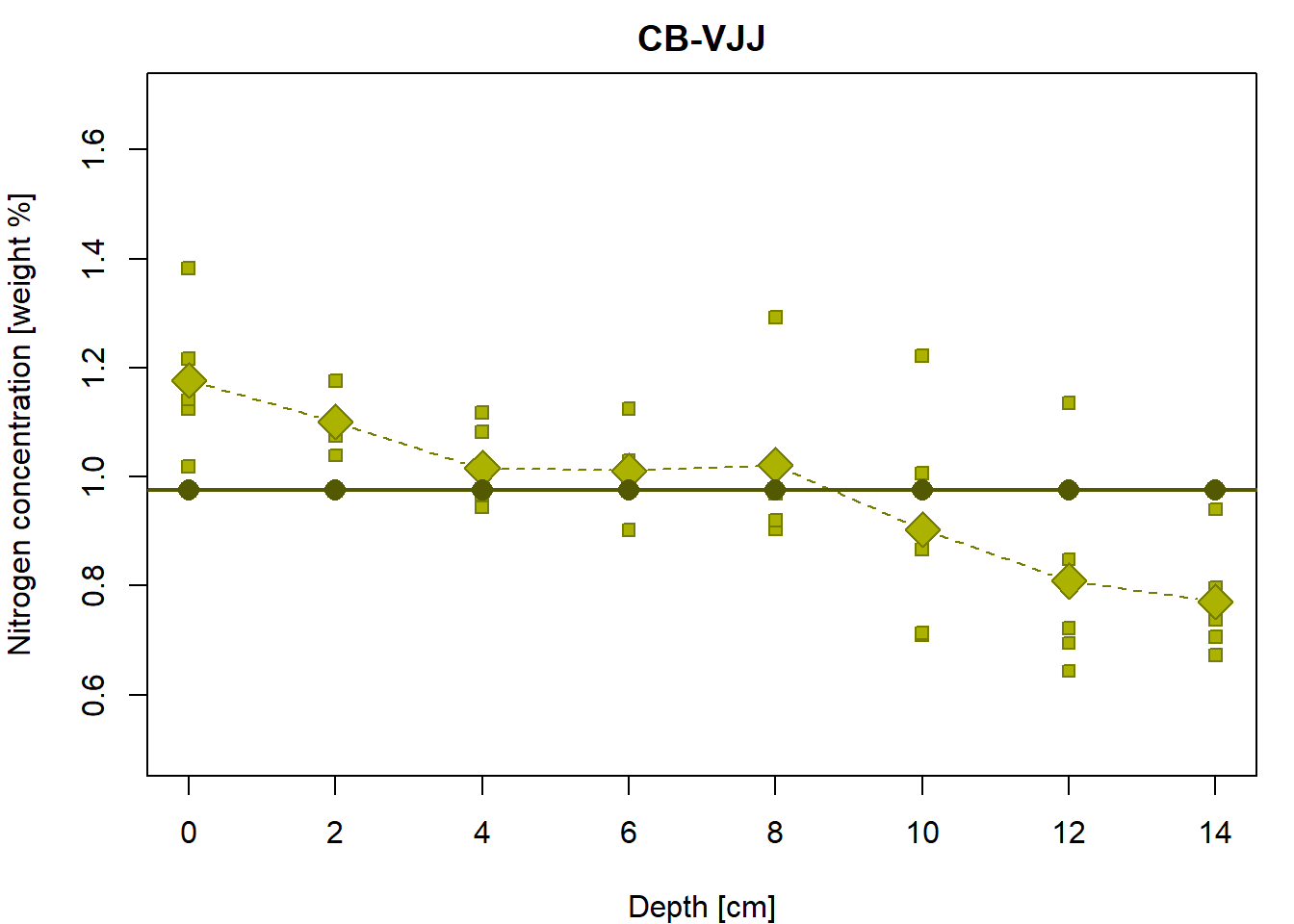
Do you see any relationship with the following?
mean(Yvalues) # beta coefficient## [1] 0.9762sd(Yvalues) # residual standard error## [1] 0.1828sd(Yvalues)/sqrt(n) # estimated sd of beta## [1] 0.0289Total sum of squares:
deviance(fit0) # SS_T (= SS_e of fit0)## [1] 1.303sum((Yvalues - mean(Yvalues))^2)## [1] 1.303Overall F-test (once more):
summary(fit1)##
## Call:
## lm(formula = N ~ depth, data = peat, subset = (group == Group))
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.1949 -0.0923 -0.0196 0.0504 0.3437
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.16907 0.03819 30.61 < 2e-16 ***
## depth -0.02755 0.00456 -6.04 5.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.132 on 38 degrees of freedom
## Multiple R-squared: 0.489, Adjusted R-squared: 0.476
## F-statistic: 36.4 on 1 and 38 DF, p-value: 5.08e-07anova(fit0, fit1)## Analysis of Variance Table
##
## Model 1: N ~ 1
## Model 2: N ~ depth
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 39 1.303
## 2 38 0.665 1 0.638 36.4 5.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Compare to standard correlation test:
cor.test(Xvalues, Yvalues)##
## Pearson's product-moment correlation
##
## data: Xvalues and Yvalues
## t = -6, df = 38, p-value = 5e-07
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.8302 -0.4963
## sample estimates:
## cor
## -0.6996Depth-as-factor model for the group “CB-VJJ”
Compare fit0 and fit1 with the model where
each depth level is considered separately:
(fit2 = lm(N ~ -1 + as.factor(depth), data = peat, subset = (group == Group)))##
## Call:
## lm(formula = N ~ -1 + as.factor(depth), data = peat, subset = (group ==
## Group))
##
## Coefficients:
## as.factor(depth)0 as.factor(depth)2 as.factor(depth)4 as.factor(depth)6 as.factor(depth)8
## 1.177 1.101 1.015 1.011 1.022
## as.factor(depth)10 as.factor(depth)12 as.factor(depth)14
## 0.904 0.809 0.771par(mfrow = c(1,1), mar = c(4,4,2,1))
# the underlying data
plot(Yvalues~Xvalues,pch = PCH[Group], col = COL[Group], bg = BGC[Group],
xlab = XLAB, ylab = YLAB, main = Group, xlim = XLIM, ylim = YLIM)
# empirical group means
lines(tabs[[g]][, "Depth"], tabs[[g]][, "Mean"], type="b",
pch = 23, cex = 2, col = COL[g], bg = BGC[g])
# regression line given by the LS estimates (for the specific values of depth)
points(subset(peat, group==Group)$depth, fitted(fit2), pch=20, cex=2, col=DARK[g])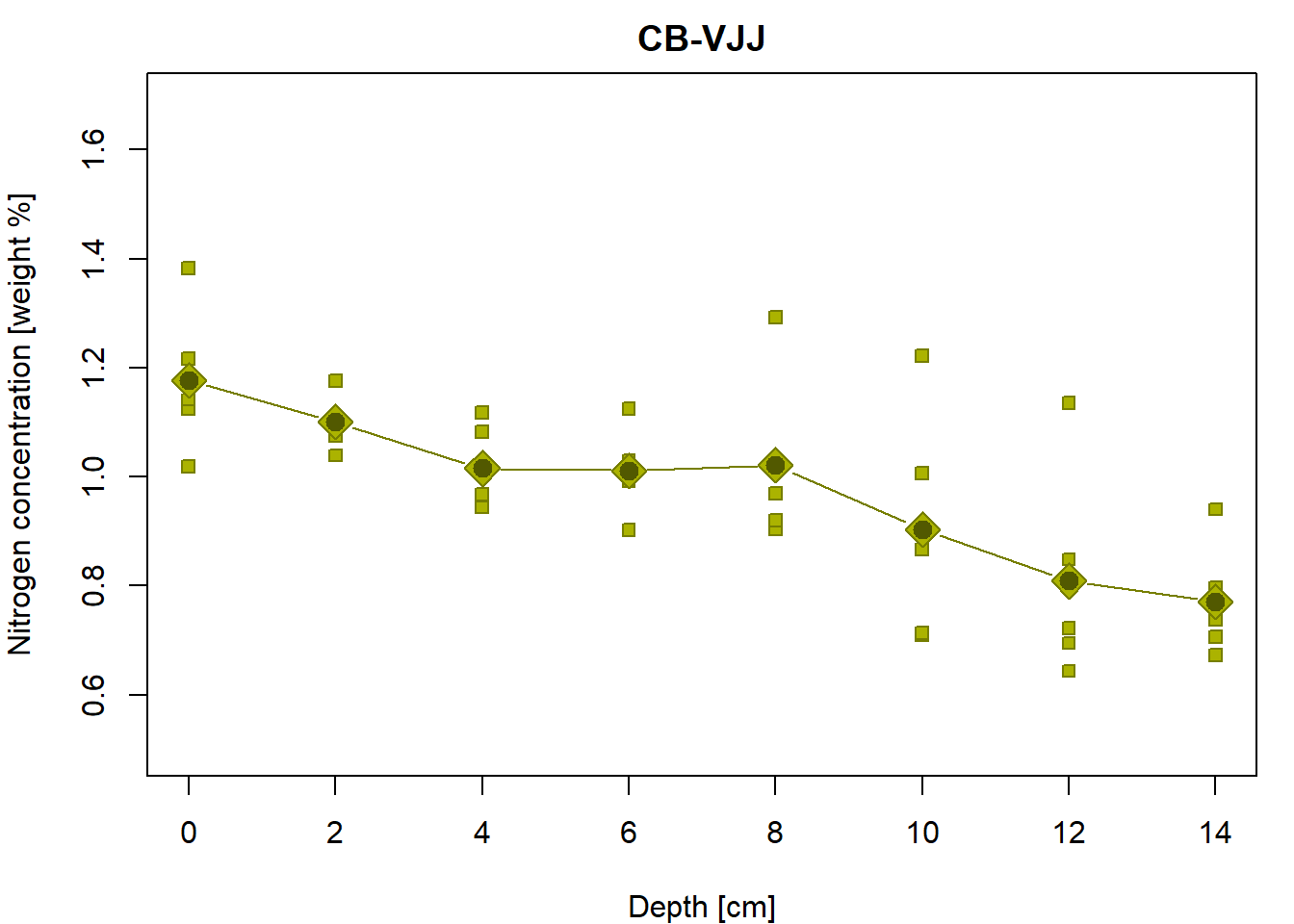
Is it possible to judge (just visually) from the third column whether one assumption of the classical linear model is satisfied? Which assumption?
print(tabs[[Group]])## Depth Mean Std.Dev n
## 1 0 1.1769 0.13512 5
## 2 2 1.1012 0.05042 5
## 3 4 1.0153 0.07849 5
## 4 6 1.0114 0.08000 5
## 5 8 1.0216 0.15842 5
## 6 10 0.9036 0.21678 5
## 7 12 0.8086 0.19743 5
## 8 14 0.7710 0.10511 5Numeric computations behind lm
We fit the simple linear regression line estimate once again with additional argument:
fit1 <- lm(N ~ depth, data = peat, subset = (group == Group), x = TRUE)
names(fit1)## [1] "coefficients" "residuals" "effects" "rank" "fitted.values" "assign"
## [7] "qr" "df.residual" "xlevels" "call" "terms" "model"
## [13] "x"The fit1 object additionally contains an element
x, the model matrix \(\mathbb{X}\):
X <- fit1[["x"]]
# X <- model.matrix(N ~ depth, peat, subset = (group == Group)) # or directly
print(X)## (Intercept) depth
## 109 1 0
## 110 1 2
## 111 1 4
## 112 1 6
## 113 1 8
## 114 1 10
## 115 1 12
## 116 1 14
## 117 1 0
## 118 1 2
## 119 1 4
## 120 1 6
## 121 1 8
## 122 1 10
## 123 1 12
## 124 1 14
## 125 1 0
## 126 1 2
## 127 1 4
## 128 1 6
## 129 1 8
## 130 1 10
## 131 1 12
## 132 1 14
## 133 1 0
## 134 1 2
## 135 1 4
## 136 1 6
## 137 1 8
## 138 1 10
## 139 1 12
## 140 1 14
## 141 1 0
## 142 1 2
## 143 1 4
## 144 1 6
## 145 1 8
## 146 1 10
## 147 1 12
## 148 1 14
## attr(,"assign")
## [1] 0 1Response \(\mathbf{Y}\) could be
obtained with y = TRUE or taken from the data:
y <- subset(peat, group == Group)[, "N"]
print(y)## [1] 1.1254 1.1048 0.9452 0.9025 0.9030 0.7088 0.6435 0.6731 1.0182 1.1100 1.0824 0.9916 1.0228 1.0068 0.8484
## [16] 0.7968 1.2168 1.0393 0.9648 1.1254 1.2924 1.2226 1.1350 0.9404 1.3827 1.1760 0.9672 1.0076 0.9694 0.8666
## [31] 0.6947 0.7064 1.1414 1.0757 1.1171 1.0298 0.9203 0.7131 0.7216 0.7384Model fitting \(\widehat{\mathbf{\beta}} = \left(\mathbb{X}^\top \mathbb{X}\right)^{-1} \mathbb{X}^\top \mathbf{Y}\):
(XtX <- t(X) %*% X) # What are the elements of XtX?## (Intercept) depth
## (Intercept) 40 280
## depth 280 2800(Xty <- t(X) %*% y) # What are the elements of Xty?## [,1]
## (Intercept) 39.05
## depth 250.19(b <- solve(XtX, Xty)) # beta## [,1]
## (Intercept) 1.16907
## depth -0.02755coef(fit1)## (Intercept) depth
## 1.16907 -0.02755Projection matrix \(\mathbb{H} = \mathbb{X} \left(\mathbb{X}^\top \mathbb{X}\right)^{-1} \mathbb{X}^\top\):
H <- X %*% solve(XtX) %*% t(X) # What is its purpose?
dim(H)## [1] 40 40all.equal(c(H%*%y),unname(fitted(fit1))) ## [1] TRUENumerically efficient way how to get it (using the QR decomposition):
Q <- qr.Q(fit1[["qr"]]) # Q matrix (orthogonal columns)
H <- qr.Q(fit1[["qr"]]) %*% t(qr.Q(fit1[["qr"]]))
dim(H)## [1] 40 40H[1:10, 1:10] # part of H matrix## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 8.333e-02 0.066667 5.000e-02 0.03333 0.01667 -6.939e-18 -0.016667 -3.333e-02 0.08333 0.066667
## [2,] 6.667e-02 0.054762 4.286e-02 0.03095 0.01905 7.143e-03 -0.004762 -1.667e-02 0.06667 0.054762
## [3,] 5.000e-02 0.042857 3.571e-02 0.02857 0.02143 1.429e-02 0.007143 -3.469e-18 0.05000 0.042857
## [4,] 3.333e-02 0.030952 2.857e-02 0.02619 0.02381 2.143e-02 0.019048 1.667e-02 0.03333 0.030952
## [5,] 1.667e-02 0.019048 2.143e-02 0.02381 0.02619 2.857e-02 0.030952 3.333e-02 0.01667 0.019048
## [6,] -6.939e-18 0.007143 1.429e-02 0.02143 0.02857 3.571e-02 0.042857 5.000e-02 0.00000 0.007143
## [7,] -1.667e-02 -0.004762 7.143e-03 0.01905 0.03095 4.286e-02 0.054762 6.667e-02 -0.01667 -0.004762
## [8,] -3.333e-02 -0.016667 -3.469e-18 0.01667 0.03333 5.000e-02 0.066667 8.333e-02 -0.03333 -0.016667
## [9,] 8.333e-02 0.066667 5.000e-02 0.03333 0.01667 0.000e+00 -0.016667 -3.333e-02 0.08333 0.066667
## [10,] 6.667e-02 0.054762 4.286e-02 0.03095 0.01905 7.143e-03 -0.004762 -1.667e-02 0.06667 0.054762max(c(abs(X %*% solve(XtX) %*% t(X) - H))) # numerically zero## [1] 6.939e-17Fitted values \(\widehat{\mathbf{Y}}\):
yhat <- H %*% y
yhat2 <- X %*% b
summary(yhat - yhat2) # Numerically only zeros## V1
## Min. :-6.66e-16
## 1st Qu.:-2.78e-16
## Median : 1.11e-16
## Mean : 1.11e-17
## 3rd Qu.: 2.50e-16
## Max. : 5.55e-16Residuals \(\mathbf{U}\):
mean(y - yhat) # Numerically zero ## [1] -7.217e-17Complement of the projection matrix \(\mathbb{M}\) - projection matrix into the space of residuals
M <- diag(nrow(H)) - H
all.equal(c(M%*%y), unname(fit1$residuals))## [1] TRUEResiduals are orthogonal to the columns of \(\mathbb{X}\), i.e. \(\mathbb{M} \cdot \mathbb{X} = \mathbb{O}\) (zero matrix):
max(abs(c(M%*%X)))## [1] 5.329e-15Therefore, since \(\mathbf{U} = \mathbb{M} \cdot \mathbf{Y}\), \[\mathbb{X}^\top \cdot \mathbf{U} = \mathbb{X}^\top \cdot \mathbb{M} \cdot \mathbf{Y} = (\mathbb{M} \cdot \mathbb{X})^\top \cdot \mathbf{Y} = \mathbf{0}.\] Because the vector of all ones \(\mathbf{1}\) is a column of \(\mathbb{X}\), also \(\mathbf{1}^\top \cdot \mathbf{U} = 0\). We obtain that the sum of residuals in a linear model with intercept must equal 0. More generally, residuals are orthogonal to the columns of \(\mathbb{X}\).
\(\mathsf{SS}_e\) (residual sum of squares):
deviance(fit1)## [1] 0.6651(SSe <- as.numeric(t(y) %*% M %*% y))## [1] 0.6651(SSe2 <- as.numeric(crossprod(y - yhat))) # The same## [1] 0.6651Residual mean square \(\mathsf{MS_e}\) (and its square root):
(df <- length(y) - r)## [1] 38(s2 <- SSe / df)## [1] 0.0175(s <- sqrt(s2))## [1] 0.1323summary(fit1)##
## Call:
## lm(formula = N ~ depth, data = peat, subset = (group == Group),
## x = TRUE)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.1949 -0.0923 -0.0196 0.0504 0.3437
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.16907 0.03819 30.61 < 2e-16 ***
## depth -0.02755 0.00456 -6.04 5.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.132 on 38 degrees of freedom
## Multiple R-squared: 0.489, Adjusted R-squared: 0.476
## F-statistic: 36.4 on 1 and 38 DF, p-value: 5.08e-07What are we going to calculate by this? What is its purpose? Can it
be found somewhere in summary(fit1)?
deviance(fit0)## [1] 1.303(SST <- as.numeric(crossprod(y - mean(y))))## [1] 1.303(WhatIsIt <- 1 - SSe / SST)## [1] 0.4895Coefficient of determination - square of the coefficient of (multiple). Also, a square of correlation between response Y and regressor X.
cor(Xvalues,Yvalues)^2 ## [1] 0.4895Variance and standard deviations of the regression coefficients’
estimates. Why is it important or useful? Can you find them somewhere in
summary(fit1)?
(varb <- s2 * solve(XtX))## (Intercept) depth
## (Intercept) 0.0014586 -1.459e-04
## depth -0.0001459 2.084e-05(sb1 <- sqrt(varb[1, 1]))## [1] 0.03819(sb2 <- sqrt(varb[2, 2]))## [1] 0.004565summary(fit1)##
## Call:
## lm(formula = N ~ depth, data = peat, subset = (group == Group),
## x = TRUE)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.1949 -0.0923 -0.0196 0.0504 0.3437
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.16907 0.03819 30.61 < 2e-16 ***
## depth -0.02755 0.00456 -6.04 5.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.132 on 38 degrees of freedom
## Multiple R-squared: 0.489, Adjusted R-squared: 0.476
## F-statistic: 36.4 on 1 and 38 DF, p-value: 5.08e-07Does the nitrogen concentration depend on the depth significantly?
(statistic <- (b[2] - 0) / sb2) ## [1] -6.036# critical value
alpha <- 0.05
(K <- qt(1 - alpha/2, df=df))## [1] 2.024# p-value
(Pval <- 2 * pt(-abs(statistic), df = df)) # Is it correct and why?## [1] 5.083e-07What is our decision? Is the test from above somewhere here?
summary(fit1) ##
## Call:
## lm(formula = N ~ depth, data = peat, subset = (group == Group),
## x = TRUE)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.1949 -0.0923 -0.0196 0.0504 0.3437
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.16907 0.03819 30.61 < 2e-16 ***
## depth -0.02755 0.00456 -6.04 5.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.132 on 38 degrees of freedom
## Multiple R-squared: 0.489, Adjusted R-squared: 0.476
## F-statistic: 36.4 on 1 and 38 DF, p-value: 5.08e-07Individual work
- Fit lines for every
groupinpeat. - Plot all the fitted lines into one plot, distinguish points and lines by color.
- You can use
jitter()to make the plot even more clear, see below.
Additional material
In some situations (especially if there are lot of data points and no reasonable insight can be made by using a standard plot) a jittered version may be used instead. In this case the depth is randomly shifted by an amount from a uniform distribution on \([-0.5,+0.5]\).
set.seed(20010911)
peat[, "jdepth"] <- peat[, "depth"] + runif(nrow(peat), -0.5, 0.5)
summary(peat)## group N depth jdepth
## CB :35 Min. :0.499 Min. : 0.00 Min. :-0.461
## CB-VJJ:40 1st Qu.:0.899 1st Qu.: 4.00 1st Qu.: 3.814
## VJJ :33 Median :1.034 Median : 8.00 Median : 7.850
## VJJ-CB:40 Mean :1.077 Mean : 7.53 Mean : 7.489
## 3rd Qu.:1.244 3rd Qu.:12.00 3rd Qu.:11.534
## Max. :1.692 Max. :14.00 Max. :14.479Another plot, this time with the jittered depth:
par(mfrow=c(1,1), mar=c(4,4,0.5,0.5))
plot(N ~ jdepth, data = peat,
xlab = XLAB, ylab = YLAB, xaxt = "n",
pch = PCH[group], col = COL[group], bg = BGC[group])
axis(1, at = seq(0, 14, by = 2))
abline(v = seq(1, 13, by = 2), col = "gray", lty = 5, lwd = 2)
legend(1.3, 1.7, legend = levels(peat[, "group"]), pch = PCH, col = COL,
pt.bg = BGC, y.intersp = 1.2)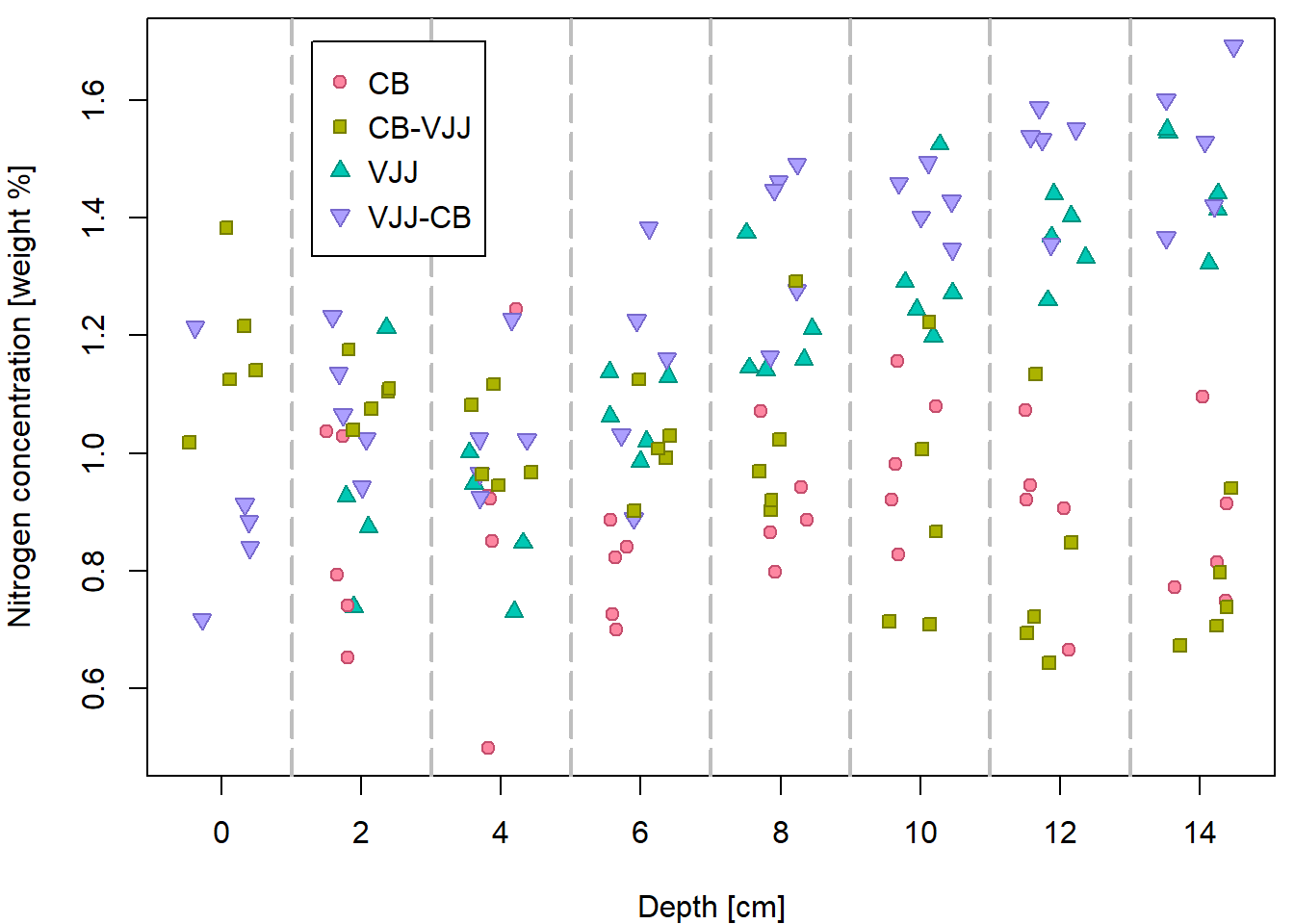
Similar plot using an R function jitter:
par(mfrow=c(1,1), mar=c(4,4,0.5,0.5))
plot(N ~ jitter(depth), data=peat,
xlab = XLAB, ylab = YLAB,
pch = PCH[group], col = COL[group], bg = BGC[group])