Simple (ordinary) linear regression model for a continuous variable
Jan Vávra
Exercise 3
Download this R markdown as: R, Rmd.
Outline of the lab session:
Simple (ordinary) linear regression model for a continuous variable \(Y \in \mathbb{R}\)
- the explanatory variable \(X \in \mathbb{R}\) is continuous,
- the explanatory variable \(X \in \mathbb{R}\) is binary,
- transformations of the explanatory variable.
Loading the data and libraries
Some necessary R packages (each package must be firstly installed in
R – this can be achieved by running the command
install.packages("package_name")). After the installation,
the libraries are initialized by
library(MASS)
library(ISLR2)The installation (command install.packages()) should be
performed just once. However, the initialization of the library – the
command library() – must be used every time when starting a
new R session.
1. Simple (ordinary) linear regression
The ISLR2 library contains the Boston data
set, which records medv (median house value) for \(506\) census tracts in Boston. We will seek
to predict medv using some of the \(17\) given predictors such as
rm (average number of rooms per house), age
(average age of houses), or lstat (percent of households
with low socioeconomic status).
head(Boston)## crim zn indus chas nox rm age dis rad tax ptratio lstat medv frm fage flstat fage2
## 1 0.00632 18 2.31 0 0.538 6.575 65.2 4.090 1 296 15.3 4.98 24.0 (6.21,6.62] TRUE (1.73,6.95] 1
## 2 0.02731 0 7.07 0 0.469 6.421 78.9 4.967 2 242 17.8 9.14 21.6 (6.21,6.62] TRUE (6.95,11.4] 1
## 3 0.02729 0 7.07 0 0.469 7.185 61.1 4.967 2 242 17.8 4.03 34.7 (6.62,8.78] TRUE (1.73,6.95] 1
## 4 0.03237 0 2.18 0 0.458 6.998 45.8 6.062 3 222 18.7 2.94 33.4 (6.62,8.78] FALSE (1.73,6.95] -1
## 5 0.06905 0 2.18 0 0.458 7.147 54.2 6.062 3 222 18.7 5.33 36.2 (6.62,8.78] TRUE (1.73,6.95] 1
## 6 0.02985 0 2.18 0 0.458 6.430 58.7 6.062 3 222 18.7 5.21 28.7 (6.21,6.62] TRUE (1.73,6.95] 1
## lstat_transformed
## 1 2.232
## 2 3.023
## 3 2.007
## 4 1.715
## 5 2.309
## 6 2.283To find out more about the data set, we can type
?Boston.
We will start with some simple explanatory analysis. For all three explanatory variables we draw a simple scatterplot.
par(mfrow = c(1,3), mar = c(4,4,0.5,0.5))
plot(medv ~ rm, data = Boston, pch = 22, bg = "gray",
ylab = "Median value [in 1000$]", xlab = "Average number of rooms")
plot(medv ~ age, data = Boston, pch = 22, bg = "gray",
ylab = "", xlab = "Proportion of owner-occupied units built prior to 1940")
plot(medv ~ lstat, data = Boston, pch = 22, bg = "gray",
ylab = "", xlab = "Lower status of the population [%].")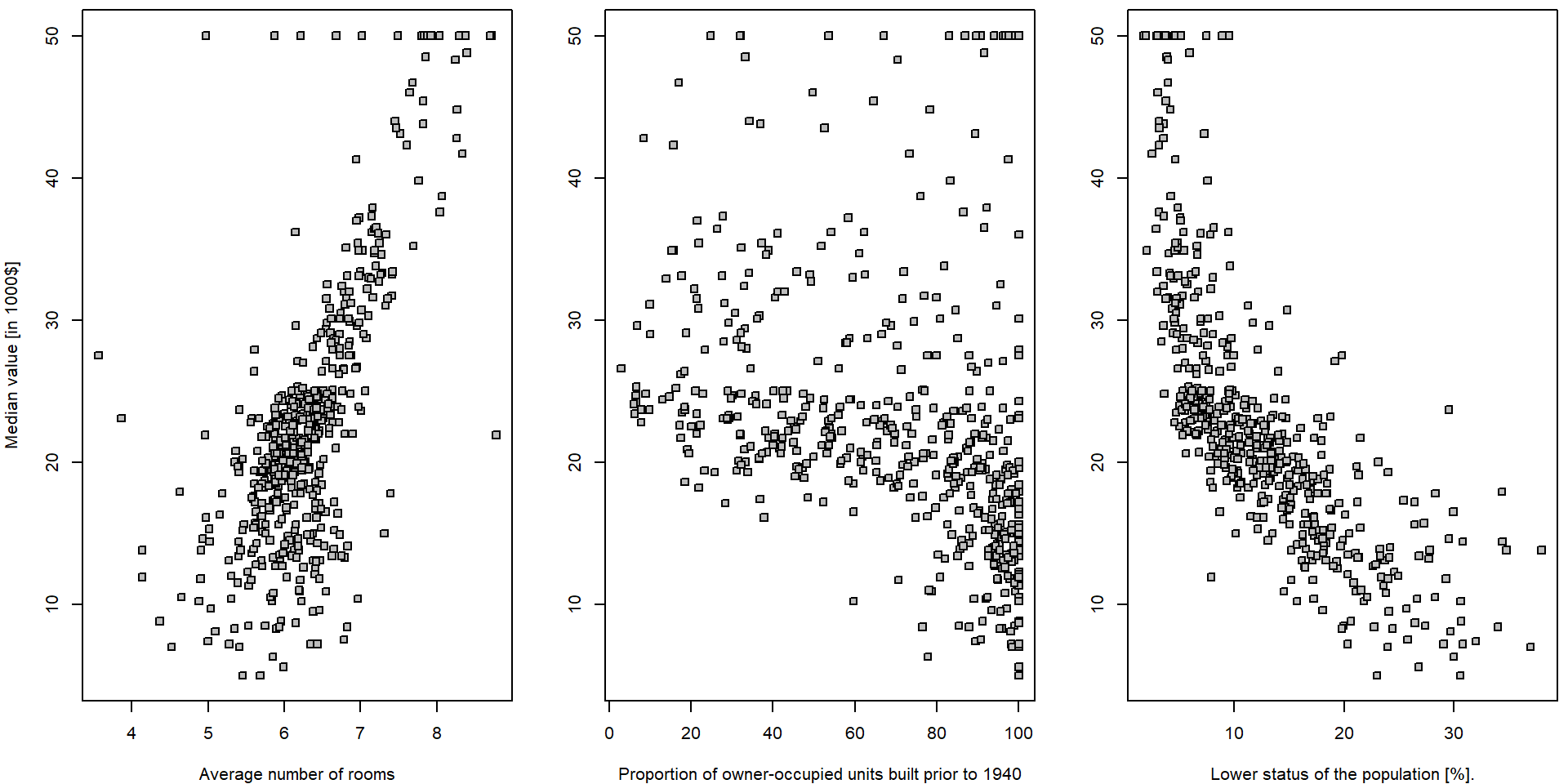
It seems that there is a positive (progressive) relationship between
the median house value (dependent variable medv) and the
average number of room in the house (independent variable
rm) and a negative relationship (regression) in terms of
the relationship between the median house value and the population
status (explanatory variable lstat). For the proportion of
the owner-occupied units build prior to 1940 (variable
age), it is not that much obvious what relationship to
expect… In all three situations we will go for a simple linear
(ordinary) regression model in terms of
\[ Y_i = a + b X_i + \varepsilon_i \qquad i = 1, \dots, 506. \]
Individual work
- For all four covariates of interest (one dependent variable and three independent variables) perform a simple exploratory analysis in terms of some estimated characteristics (e.g., the estimate for mean, median, variance, standard error).
- Use some graphical tools to visualize the structure in the given data – the four covariates of interest (e.g. some simple boxplots).
- Perform a more detailed exploratory analysis in terms of splitting
the data into some reasonable subgroups and estimated the corresponding
characteristics in each subgroup separately (for instance, you can
consider two subgroups by distinguishing the proportion
agebelow 50% and above 50%).
vars <- c("medv", "rm", "age", "lstat")
summary(Boston[,vars])## medv rm age lstat
## Min. : 5.0 Min. :3.56 Min. : 2.9 Min. : 1.73
## 1st Qu.:17.0 1st Qu.:5.89 1st Qu.: 45.0 1st Qu.: 6.95
## Median :21.2 Median :6.21 Median : 77.5 Median :11.36
## Mean :22.5 Mean :6.29 Mean : 68.6 Mean :12.65
## 3rd Qu.:25.0 3rd Qu.:6.62 3rd Qu.: 94.1 3rd Qu.:16.95
## Max. :50.0 Max. :8.78 Max. :100.0 Max. :37.97apply(Boston[,vars], 2, sd)## medv rm age lstat
## 9.1971 0.7026 28.1489 7.1411cor(Boston[,vars])## medv rm age lstat
## medv 1.0000 0.6954 -0.3770 -0.7377
## rm 0.6954 1.0000 -0.2403 -0.6138
## age -0.3770 -0.2403 1.0000 0.6023
## lstat -0.7377 -0.6138 0.6023 1.0000for(x in vars[2:4]){
Boston[,paste0("f",x)] <- cut(Boston[,x], breaks = quantile(Boston[,x], seq(0,1, length.out = 5)))
}
fxs <- paste0("f",vars[2:4])
summary(Boston[,fxs])## frm fage flstat
## (3.56,5.89]:126 (2.9,45] :126 (1.73,6.95]:126
## (5.89,6.21]:126 (45,77.5] :126 (6.95,11.4]:126
## (6.21,6.62]:126 (77.5,94.1]:126 (11.4,17] :126
## (6.62,8.78]:127 (94.1,100] :127 (17,38] :127
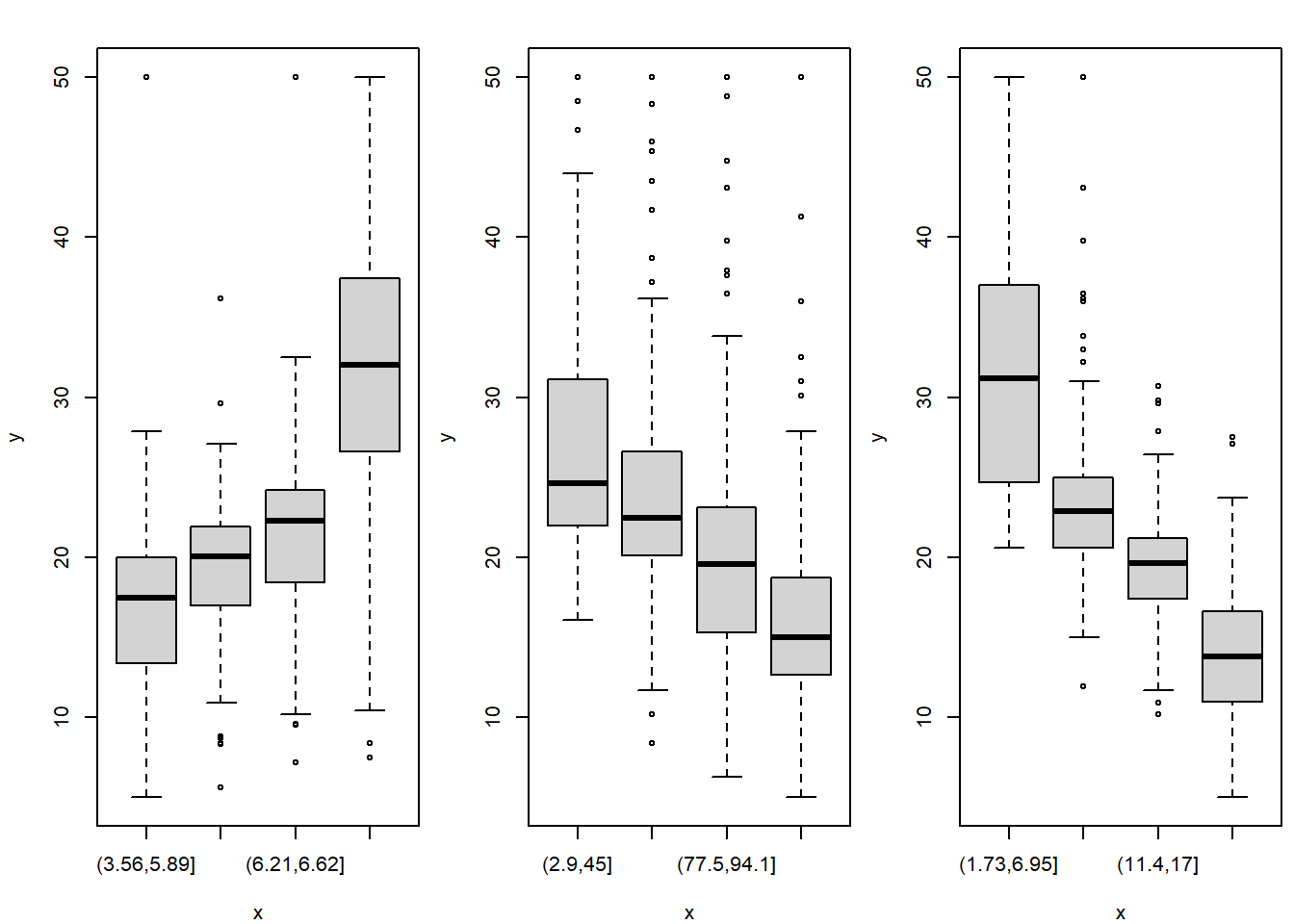
## NA's : 1 NA's : 1 NA's : 1par(mfrow = c(1,3), mar = c(4,4,2,0.5))
for(x in fxs){
plot(x = Boston[,x], y = Boston$medv)
}
In order to get some insight into the relationship \(Y = f(X) + \varepsilon\) (and to judge the
appropriateness of the linear line to be used as a surrogate for \(f(\cdot)\)) we can take a look at some
(non-parametric) data smoothing – function lowess() in
R:
par(mfrow = c(1,3), mar = c(4,4,0.5,0.5))
plot(medv ~ rm, data = Boston, pch = 22, bg = "gray",
ylab = "Median value [in 1000$]", xlab = "Average number of rooms")
lines(lowess(Boston$medv ~ Boston$rm, f = 2/3), col = "red", lwd = 2)
plot(medv ~ age, data = Boston, pch = 22, bg = "gray",
ylab = "", xlab = "Proportion of owner-occupied units built prior to 1940")
lines(lowess(Boston$medv ~ Boston$age, f = 2/3), col = "red", lwd = 2)
plot(medv ~ lstat, data = Boston, pch = 22, bg = "gray",
ylab = "", xlab = "Lower status of the population [%].")
lines(lowess(Boston$medv ~ Boston$lstat, f = 2/3), col = "red", lwd = 2)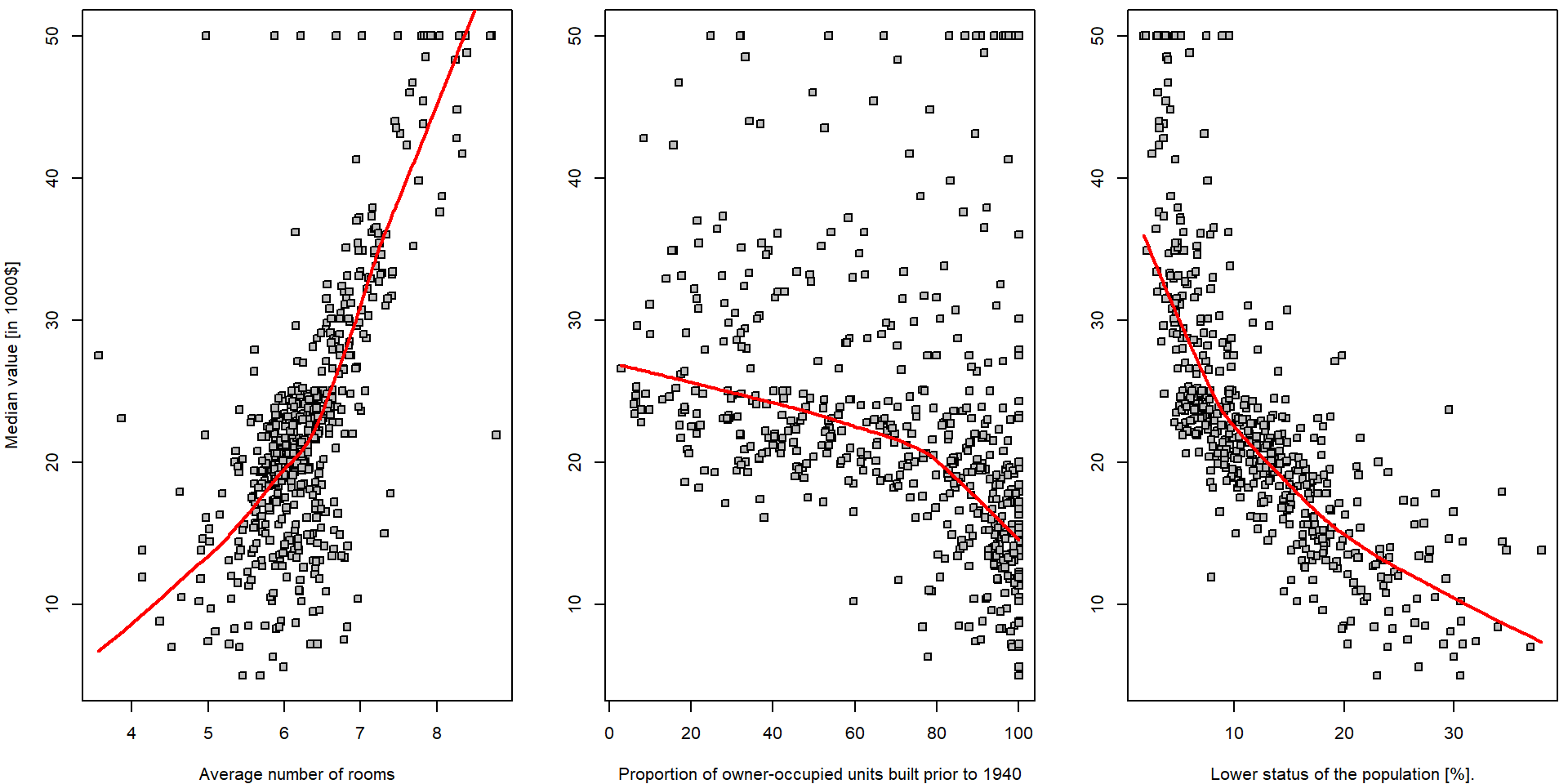
Note, that there is hardly any reasonable analytical expression for
the red curves above (the specific form of the function \(f(\cdot)\)). Also note the parameter
f = 2/3 in the function lowess(). Run the same
R code with different options for the value of this parameter to see
differences.
Explanatory variable lstat
We will now fit a simple linear regression line through the data
using the R function lm(). In the implementation used
below, the analytical form of the function \(f(\cdot)\) being used to “smooth” the data
is \[
f(x) = a + bx
\] for some intercept parameter \(a \in
\mathbb{R}\) and the slope parameter \(b \in \mathbb{R}\). The following R code
fits a simple linear regression line, with medv as the
response (depedent variable) and lstat as the predictor
(explanatory/indepenent variable/covariate). The basic syntax is
lm(y ~ x, data), where y is the response,
x is the predictor, and data is the data set
in which these two variables are kept.
lm.fit <- lm(medv ~ lstat, data = Boston)If we type lm.fit, some basic information about the
model is output. For more detailed information, we use
summary(lm.fit). This gives us \(p\)-values and standard errors for the
coefficients, as well as the \(R^2\)
statistic and \(F\)-statistic for the
model.
lm.fit##
## Call:
## lm(formula = medv ~ lstat, data = Boston)
##
## Coefficients:
## (Intercept) lstat
## 34.55 -0.95This fits a straight line through the data (the line intersects the \(y\) axis at the point \(34.55\) and the value of the slope parameter – which equals \(-0.95\) – means that for each unit increase on the \(x\) axis the line drops by \(0.95\) units with respect to the \(y\) axis).
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(medv ~ lstat, data = Boston, pch = 22, bg = "gray",
ylab = "Median value [in 1000$]", xlab = "Lower status of the population [%].")
lines(lowess(Boston$medv ~ Boston$lstat, f = 2/3), col = "red", lwd = 2)
abline(lm.fit, col = "blue", lwd = 2)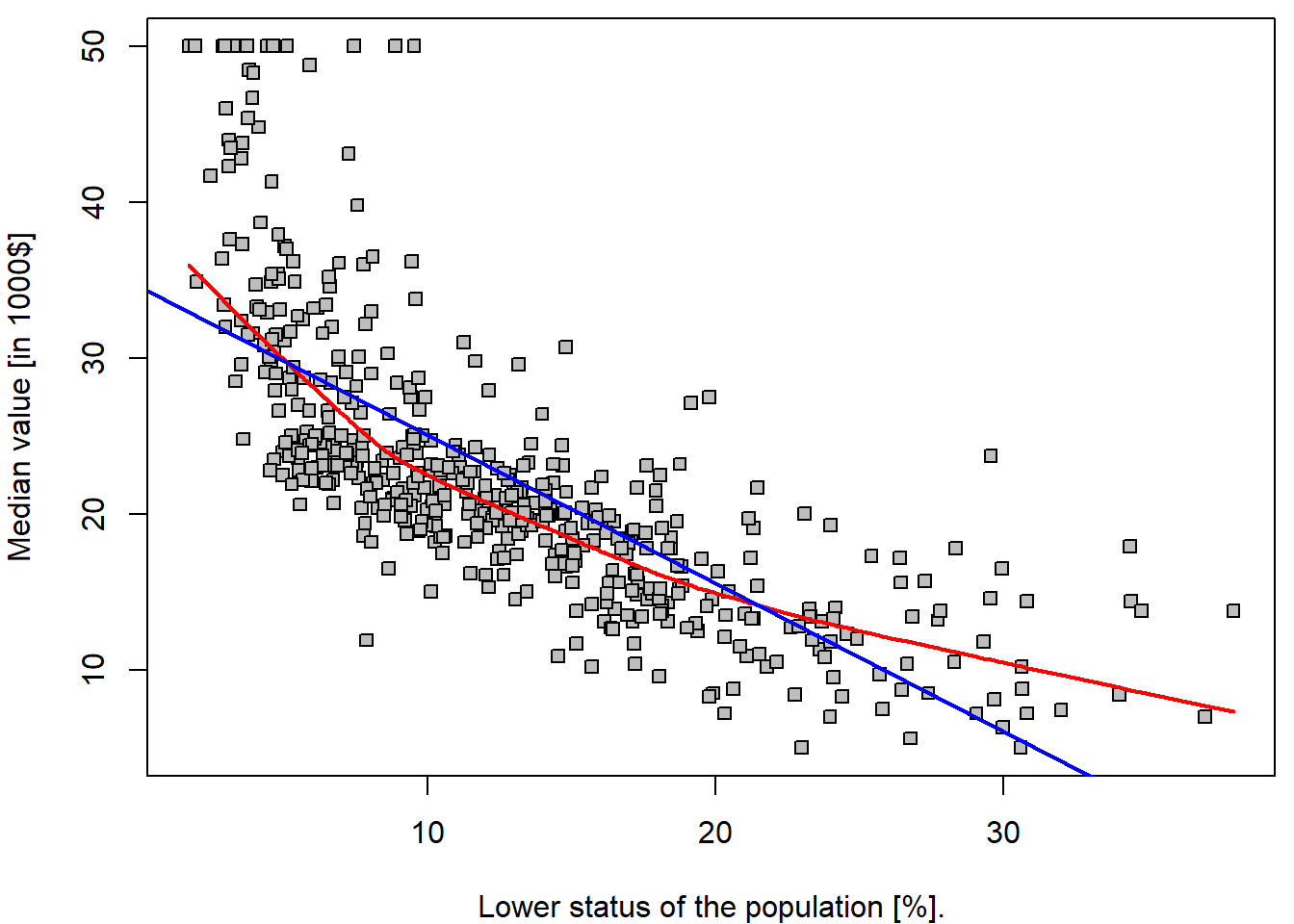
It is important to realize that the estimated parameters \(34.55\) and \(-0.95\) are given realization of two random variables – \(\widehat{a}\) and \(\widehat{b}\) (i.e., for another dataset, or another subset of the observations we would obtained different values of the estimated parameters). Thus, we are speaking about random quantities here and, therefore, it is reasonable to inspect some of their statistical properties (such as the corresponding mean parameters, variance of the estimates, or even their full distribution…).
Some of the statistical properties (i.e., the empirical estimates for the standard errors of the estimates) can be obtained by the following command:
(sum.lm.fit <- summary(lm.fit))##
## Call:
## lm(formula = medv ~ lstat, data = Boston)
##
## Residuals:
## Min 1Q Median 3Q Max
## -15.17 -3.99 -1.32 2.03 24.50
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 34.5538 0.5626 61.4 <2e-16 ***
## lstat -0.9500 0.0387 -24.5 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6.22 on 504 degrees of freedom
## Multiple R-squared: 0.544, Adjusted R-squared: 0.543
## F-statistic: 602 on 1 and 504 DF, p-value: <2e-16We can use the names() function in order to find out
what other pieces of information are stored in lm.fit.
Although we can extract these quantities by
name—e.g. lm.fit$coefficients—it is safer to use the
extractor functions like coef() to access them.
names(lm.fit)## [1] "coefficients" "residuals" "effects" "rank" "fitted.values" "assign"
## [7] "qr" "df.residual" "xlevels" "call" "terms" "model"coef(lm.fit)## (Intercept) lstat
## 34.55 -0.95It is important to realize, that the estimates \(\widehat{a}\) and \(\widehat{b}\) are random variables. They have the corresponding variances \(\mathsf{var} (\widehat{a})\) and \(\mathsf{var} (\widehat{b})\) but the values that we see in the table above are just the estimates of this theoretical quantities – thus, mathematically correctly, we should use the notation \[ \widehat{\mathsf{var}(\widehat{a})} \approx 0.5626^2 \qquad\text{and}\qquad \widehat{\mathsf{var}(\widehat{b})} \approx 0.03873^2 \] The values in the table (second column) are the estimates for the true standard errors of \(\widehat{a}\) and \(\widehat{b}\) (because we do not now the true variance, or standard error respectively, of the error terms \(\varepsilon_i\) in the underlying model \(Y_i = a + b X_i + \varepsilon_i\), where we only assume that \(\varepsilon_i \sim (0, \sigma^2)\)).
The unknown variance \(\sigma^2 >
0\) is also estimated in the output above – see the value for the
Residual standard error. Thus, we have \(\widehat{\sigma^2} = 6.216^2\).
### model residuals -- estimates for random errors and their estimated variance/standard error
var(lm.fit$residuals)## [1] 38.56sqrt(var(lm.fit$residuals)) # not exactly as in summary, var divides by (n-1) not (n-rank) ## [1] 6.21The intercept parameter \(a \in
\mathbb{R}\) and the slope parameter \(b \in \mathbb{R}\) are unknown but fixed
parameters and we have the corresponding (point) estimates for both –
random quantities \(\widehat{a}\) and
\(\widehat{b}\). Thus, it is also
reasonable to ask for an interval estimates instead – the confidence
intervals for \(a\) and \(b\).
This can be obtained by the command confint().
confint(lm.fit)## 2.5 % 97.5 %
## (Intercept) 33.448 35.659
## lstat -1.026 -0.874The estimated parameters \(\widehat{a}\) and \(\widehat{b}\) can be used to estimate some
characteristic of the distribution of the dependent variable \(Y\) (i.e., medv) given the
value of the independent variable “\(X =
x\)” (i.e., lstat). In other words, the estimated
parameters can be used to estimate the conditional expectation of the
conditional distribution of \(Y\) given
“\(X = x\)”, respectively \[
\widehat{\mu_x} = \widehat{E[Y | X = x]} = \widehat{a} + \widehat{b}x.
\]
However, sometimes it can be also useful to “predict” the value of \(Y\) for a specific value of \(X\). As far as \(Y\) is random (even conditionally on \(X = x\)) we need to give some characteristic of the whole conditional distribution – and this characteristic is said to be a prediction for \(Y\). It is common to use the conditional expectation for this purpose.
In the R program, we can use the predict() function,
which also produce the confidence intervals and prediction intervals for
the prediction of medv for a given value of
lstat.
(pred.conf <- predict(lm.fit, data.frame(lstat = (c(5, 10, 15))), interval = "confidence"))## fit lwr upr
## 1 29.80 29.01 30.60
## 2 25.05 24.47 25.63
## 3 20.30 19.73 20.87(pred.pred <- predict(lm.fit, data.frame(lstat = (c(5, 10, 15))), interval = "prediction"))## fit lwr upr
## 1 29.80 17.566 42.04
## 2 25.05 12.828 37.28
## 3 20.30 8.078 32.53For instance, the 95,% confidence interval associated with a
lstat value of 10 is \((24.47,
25.63)\), and the 95,% prediction interval is \((12.83, 37.28)\). As expected, the
confidence and prediction intervals are centered around the same point
(a predicted value of \(25.05\) for
medv when lstat equals 10), but the latter are
substantially wider. Can you explain why?
Individual work
- Add confidence and prediction intervals to the fitted line.
grid <- 0:100
pred_conf <- predict(lm.fit, newdata = data.frame(lstat = grid), interval = "confidence")
pred_pred <- predict(lm.fit, newdata = data.frame(lstat = grid), interval = "predict")
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
plot(medv ~ lstat, data = Boston, pch = 22, bg = "gray",
ylab = "Median value [in 1000$]", xlab = "Lower status of the population [%].")
lines(lowess(Boston$medv ~ Boston$lstat, f = 2/3), col = "red", lwd = 2)
abline(lm.fit, col = "blue", lwd = 2)
lines(x = grid, y = pred_conf[,"lwr"], col = "darkviolet", lty = 2, lwd = 2)
lines(x = grid, y = pred_conf[,"upr"], col = "darkviolet", lty = 2, lwd = 2)
lines(x = grid, y = pred_pred[,"lwr"], col = "chocolate", lty = 3, lwd = 2)
lines(x = grid, y = pred_pred[,"upr"], col = "chocolate", lty = 3, lwd = 2)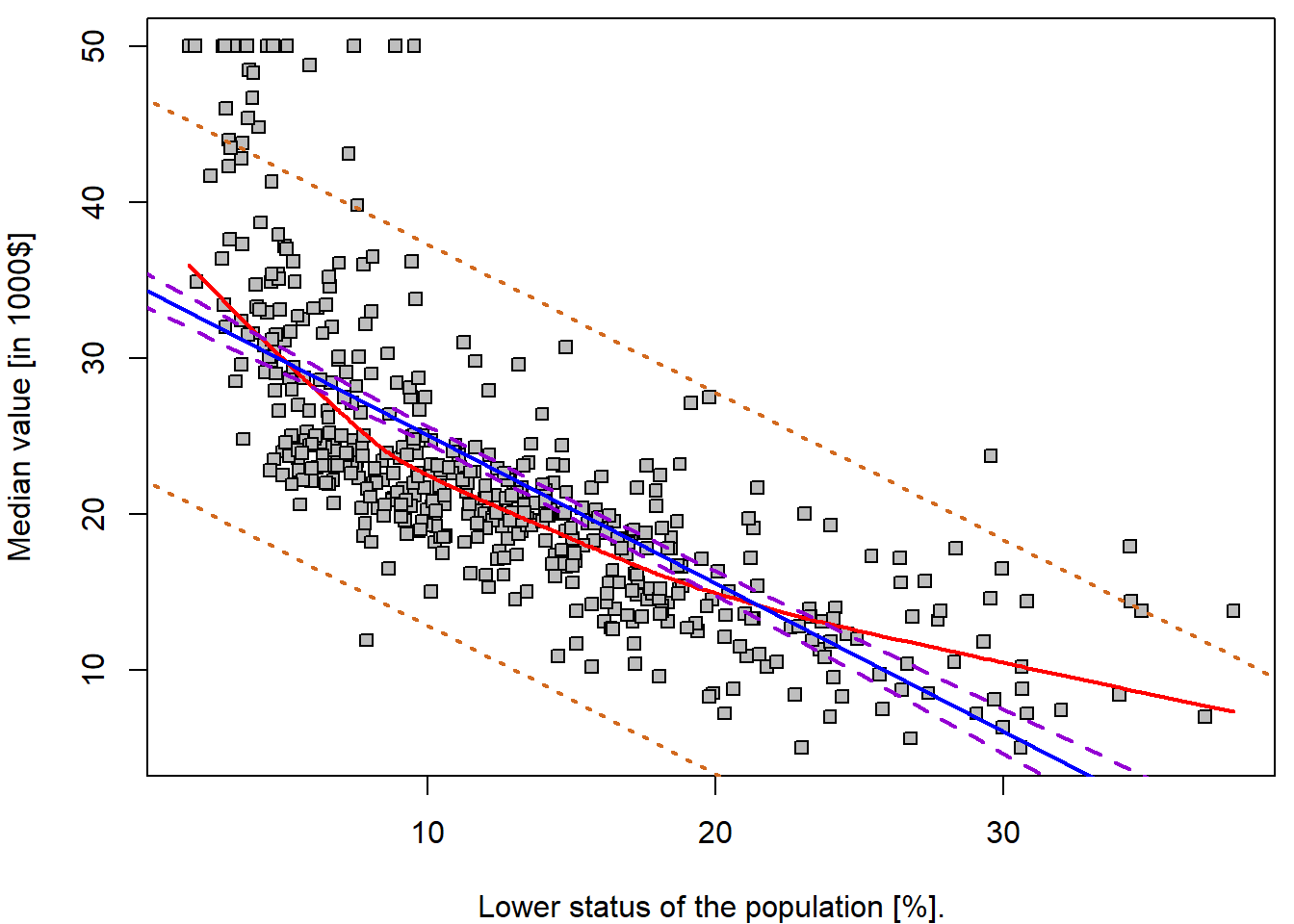
Some “goodness-of-fit” diagnostics
Next we examine some diagnostic plots – but we will discuss different diagnostics tools later.
Four diagnostic plots are automatically produced by applying the
plot() function directly to the output from
lm(). In general, this command will produce one plot at a
time, and hitting Enter will generate the next plot. However,
it is often convenient to view all four plots together. We can achieve
this by using the par() and mfrow() functions,
which tell R to split the display screen into separate
panels so that multiple plots can be viewed simultaneously. For example,
par(mfrow = c(2, 2)) divides the plotting region into a
\(2 \times 2\) grid of panels:
par(mfrow = c(2, 2), mar = c(4,4,2,0.5))
plot(lm.fit)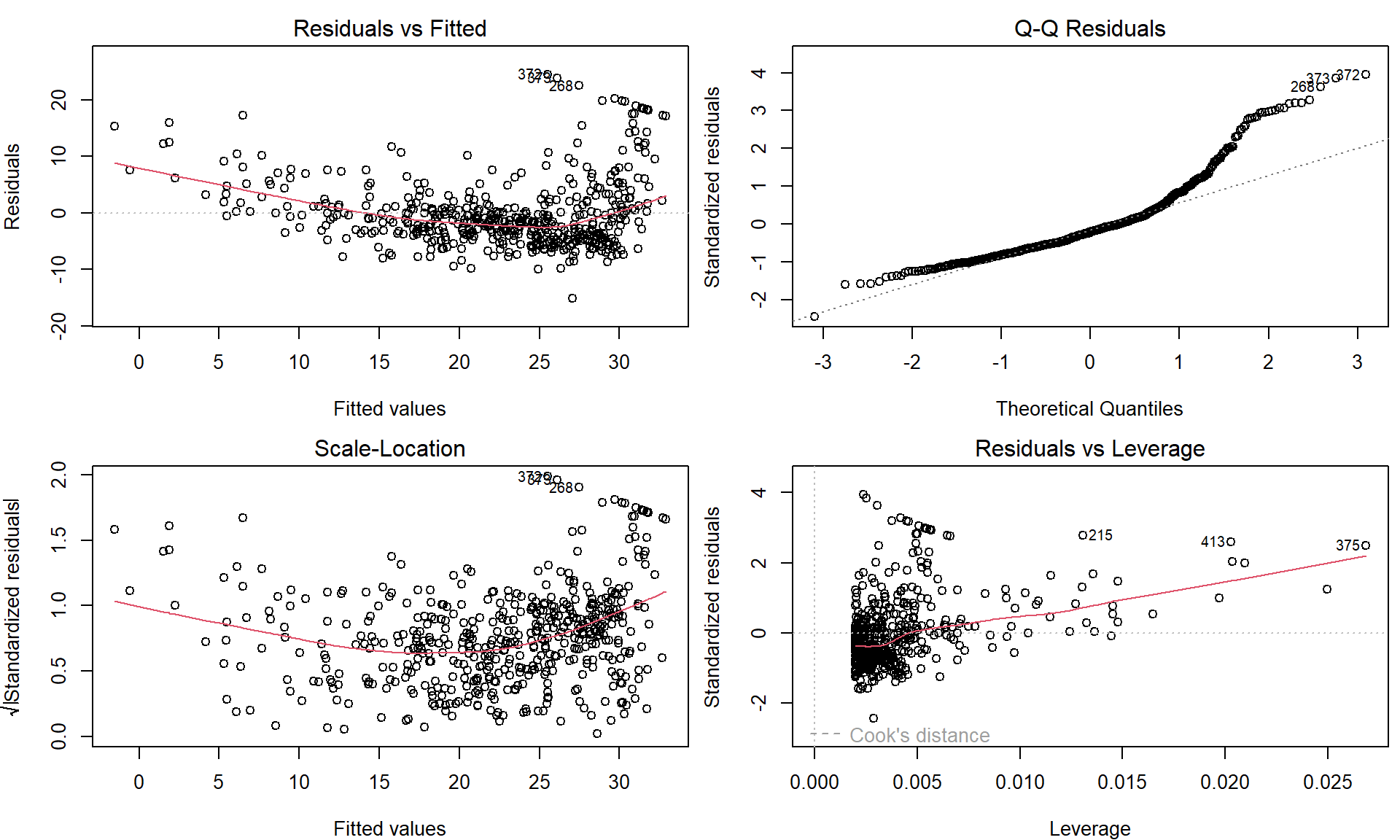
These plots can be used to judge the quality of the model that was used – in our particular case we used a simple linear regression line of the form \(Y = a + bX + \varepsilon\).
Individual work
- Repeat the previous R code and fit simple linear regression models
also for the remaining explanatory variable mentioned at the beginning –
ageandrm. - Compare the quality of all three models – visually by looking at the scatterplot and the fitted line and, also, by looking at four diagnostic plots as shown above.
- Which of the three models is the one that you consider the most reliable? Which one do you consider mostly unreliable?
- Is there any relationship/expectation with respect to the estimated covariances and correlations below?
cov(Boston$medv, Boston$rm)## [1] 4.493cov(Boston$medv, Boston$age)## [1] -97.59cov(Boston$medv, Boston$lstat)## [1] -48.45cor(Boston$medv, Boston$rm)## [1] 0.6954cor(Boston$medv, Boston$age)## [1] -0.377cor(Boston$medv, Boston$lstat)## [1] -0.73772. Binary explanatory variable \(X\)
Until now, the explanatory variable \(X\) was treated as a continuous variable. However, this variable can be also used as a binary information (and also as a categorical variable, but this will be discussed later).
We already mentioned a situation where the proportion of the owner-occupied houses is below 50% and above. Thus, we will create another variable in the original data that will reflect this information.
Boston$fage <- (Boston$age > 50)
table(Boston$fage)##
## FALSE TRUE
## 147 359We can look at both subpopulations by the means of two boxplots for instance:
par(mfrow = c(1,1), mar = c(4,4,0.5,0.5))
boxplot(medv ~ fage, data = Boston, col = "lightblue",
xlab = "Proportion of owner-occupied houses prior to 1940 is above 50%", ylab = "Median house value")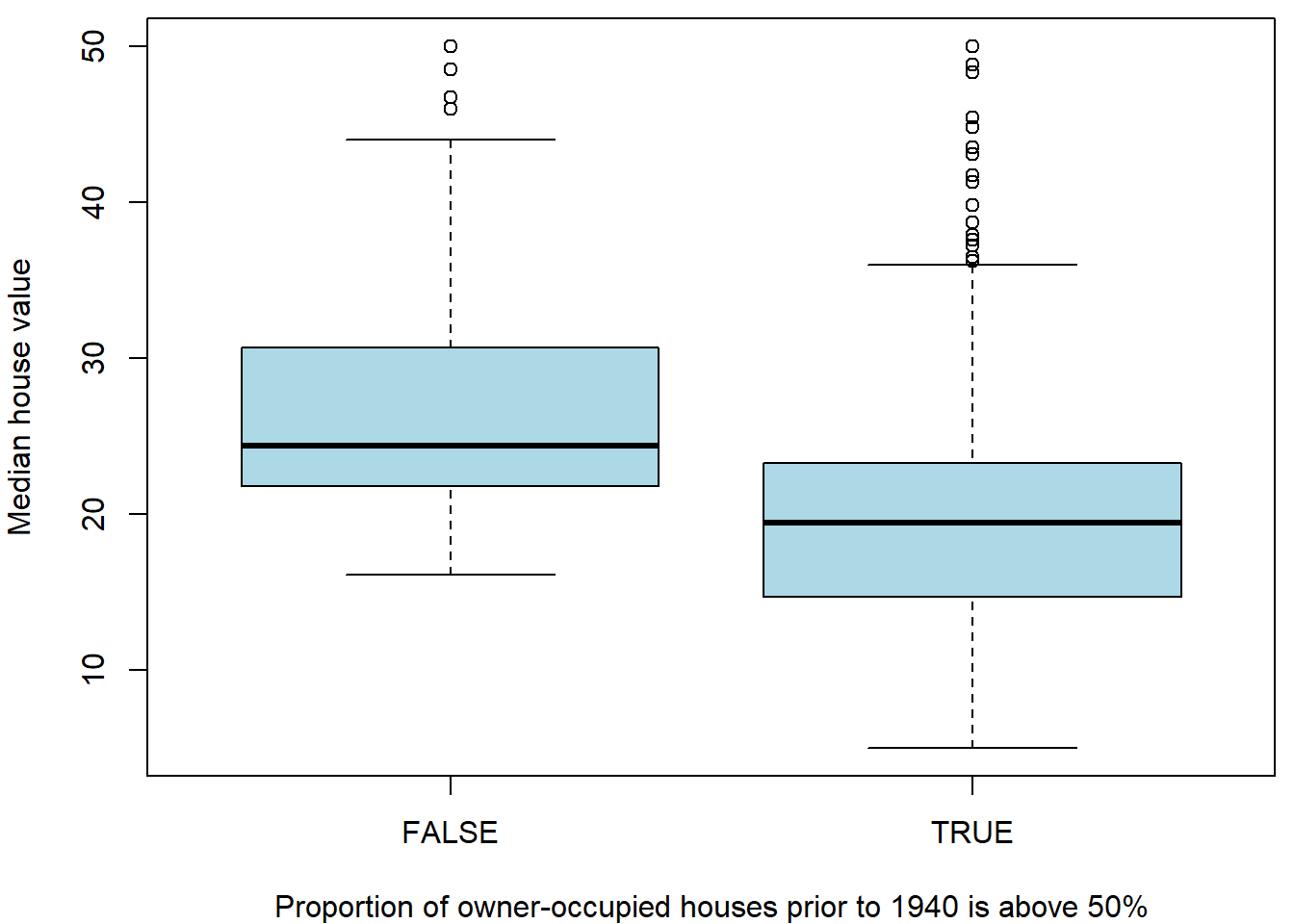
The figure above somehow corresponds with the scatterplot of
medv against age where we observed that higher
proportion (i.e., higher values of the age variable) are
associated with rather lower median house values (dependent variable
\(Y \equiv\) medv). In the
boxplot above, we can also see that higher proportions of the
owner-occupied houses prior to 1940 (the sub-population where the
proportion is above \(50\%\)) are
associated with rather lower median house values.
What will happen when this information (explanatory variable
fage which only takes two values – one for true and zero
otherwise) will be used in a simple linear regression model?
lm.fit2 <- lm(medv ~ fage, data = Boston)
lm.fit2##
## Call:
## lm(formula = medv ~ fage, data = Boston)
##
## Coefficients:
## (Intercept) fageTRUE
## 26.69 -5.86And the correponding statistical summary of the model:
summary(lm.fit2)##
## Call:
## lm(formula = medv ~ fage, data = Boston)
##
## Residuals:
## Min 1Q Median 3Q Max
## -15.83 -5.72 -1.73 2.90 29.17
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 26.693 0.727 36.7 <2e-16 ***
## fageTRUE -5.864 0.863 -6.8 3e-11 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.81 on 504 degrees of freedom
## Multiple R-squared: 0.084, Adjusted R-squared: 0.0821
## F-statistic: 46.2 on 1 and 504 DF, p-value: 3.04e-11Can you see some analogy with the following (partial) exploratory characteristic (respectively their sample estimates)?
mean(Boston$medv[Boston$fage == F])## [1] 26.69mean(Boston$medv[Boston$fage == T])## [1] 20.83mean(Boston$medv[Boston$fage == T]) - mean(Boston$medv[Boston$fage == F])## [1] -5.864The estimated model in this case takes a form \[ Y = a + bX + \varepsilon \] where \(x\) can only take two specific values – it is either equal to zero and, thus, the model becomes \(f(x) = a\) or the value of \(x\) is equal to 1 and the model becomes \(f(x) = a + b\). Note, that any other parametrization of the explanatory variable \(X\) (for instance, \(x = -1\) for the subpopulation with the proportion below 50% and \(x = 1\) for the subpopulation with the proportion above 50%) gives mathematically equivalent model.
### different parametrization of two subpopulations (using +/- 1 instead of 0/1)
Boston$fage2 <- (2 * as.numeric(Boston$fage) - 1)
head(Boston)## crim zn indus chas nox rm age dis rad tax ptratio lstat medv frm fage flstat fage2
## 1 0.00632 18 2.31 0 0.538 6.575 65.2 4.090 1 296 15.3 4.98 24.0 (6.21,6.62] TRUE (1.73,6.95] 1
## 2 0.02731 0 7.07 0 0.469 6.421 78.9 4.967 2 242 17.8 9.14 21.6 (6.21,6.62] TRUE (6.95,11.4] 1
## 3 0.02729 0 7.07 0 0.469 7.185 61.1 4.967 2 242 17.8 4.03 34.7 (6.62,8.78] TRUE (1.73,6.95] 1
## 4 0.03237 0 2.18 0 0.458 6.998 45.8 6.062 3 222 18.7 2.94 33.4 (6.62,8.78] FALSE (1.73,6.95] -1
## 5 0.06905 0 2.18 0 0.458 7.147 54.2 6.062 3 222 18.7 5.33 36.2 (6.62,8.78] TRUE (1.73,6.95] 1
## 6 0.02985 0 2.18 0 0.458 6.430 58.7 6.062 3 222 18.7 5.21 28.7 (6.21,6.62] TRUE (1.73,6.95] 1
## lstat_transformed
## 1 2.232
## 2 3.023
## 3 2.007
## 4 1.715
## 5 2.309
## 6 2.283lm.fit3 <- lm(medv ~ fage2, data = Boston)
lm.fit3##
## Call:
## lm(formula = medv ~ fage2, data = Boston)
##
## Coefficients:
## (Intercept) fage2
## 23.76 -2.93Individual work
Usually, with different parametrization we obtain different
interpretation options and statistical characteristics for different
quantities when calling the function summary().
- What is the interpretation of the estimates \(\widehat{a}\) and \(\widehat{b}\) in this case?
- Recall, that the model \(f(x) = a + bx\) for \(x = \pm 1\) takes now the form \(f(x) = a - b\) if the proportion is below \(50\%\) and the model becomes \(f(x) = a + b\) if the proportion is above \(50\%\).
- Can you reconstruct both estimates for subpopulations means using the model above?
- Try to think of some other parametrization of the independent variable \(X\) (the information whether the proportion is above or below \(50\%\)).
- In the model above we have the estimate for the intercept parameter \(\widehat{a} = 23.76\). Can you guess what does it stands for? What theoretical characteristic does it estimate?
(mean(Boston$medv[Boston$fage2 == -1]) + mean(Boston$medv[Boston$fage2 == 1]))/2## [1] 23.76- Recall, that two subpopulations are not equal with respect to their size. What would be estimated by the intercept parameter estimate \(\widehat{a}\) if both subpopulations would be equal with respect to the size (i.e., balanced data)?
- Compare
summary()outputs for the two models. Which quantities are the same for both parametrizations? - How would you fit a model where coefficients correspond to individual groups (no intercept)?
3. Transformations of the predictor variable \(X\)
We already mentioned some transformation (parametrizations) of the independent/exploratory variable \(X\) in a situation where \(X\) stands for a binary information (yes/no, true/false, \(\pm 1\), \(0/1\), etc.). However, different transformations can be also used for the independent covariate \(X\) if it stands for a continuous variable.
Consider the first model
lm.fit##
## Call:
## lm(formula = medv ~ lstat, data = Boston)
##
## Coefficients:
## (Intercept) lstat
## 34.55 -0.95and the model
lm(medv ~ I(lstat/100), data = Boston)##
## Call:
## lm(formula = medv ~ I(lstat/100), data = Boston)
##
## Coefficients:
## (Intercept) I(lstat/100)
## 34.6 -95.0What type of transformation is used in this example and what is the
underlying interpretation of this different parametrization of \(X\)? Such and similar parametrization are
typically used to improve the interpretation of the final model. For
instance, using the first model lm.fit the interpretation
of the estimated intercept parametr \(\widehat{a}\) can be given as follows:
“The estimated expected value for the median house value is 34.5
thousand dollars if the lower population status is at the zero
level.” But this may not be realistic when looking at the data –
indeed, the minimum observed value for lstat is \(1.73\). Thus, it can be more suitable to
use a different interpretation of the intercept parameter. Consider the
following model and the summary statistics for lstat:
lm(medv ~ I(lstat - 11.36), data = Boston)##
## Call:
## lm(formula = medv ~ I(lstat - 11.36), data = Boston)
##
## Coefficients:
## (Intercept) I(lstat - 11.36)
## 23.76 -0.95summary(Boston$lstat)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.73 6.95 11.36 12.65 16.96 37.97What is the interpretation of the intercept parameter estimate in this case? Can you obtain the value of the estimate using the original model?
The transformations of the independent covariates can be, however, also used to improve the model fit. For instance, consider the following model
Boston$lstat_transformed <- sqrt(Boston$lstat)
lm.fit4 <- lm(medv ~ lstat_transformed, data = Boston)Both model can be compared visually:
par(mfrow = c(1,3), mar = c(4,4,2,0.5))
plot(medv ~ lstat, data = Boston, pch = 22, bg = "gray",
ylab = "Median value [in 1000$]", xlab = "Lower status of the population [%].",
main = "Original scale X - linear trend")
abline(lm.fit, col = "blue", lwd = 2)
plot(medv ~ lstat_transformed, data = Boston, pch = 22, bg = "gray",
ylab = "", xlab = "Lower status of the population squared [sqrt(%)].",
main = "Square root of X")
abline(lm.fit4, col = "blue", lwd = 2)
plot(medv ~ lstat, data = Boston, pch = 22, bg = "gray",
ylab = "", xlab = "Lower status of the population [%].",
main = "Sqrt X fit on original scale")
xGrid <- seq(0,40, length = 1000)
yValues <- lm.fit4$coeff[1] + lm.fit4$coeff[2] * sqrt(xGrid)
lines(yValues ~ xGrid, col = "blue", lwd = 2)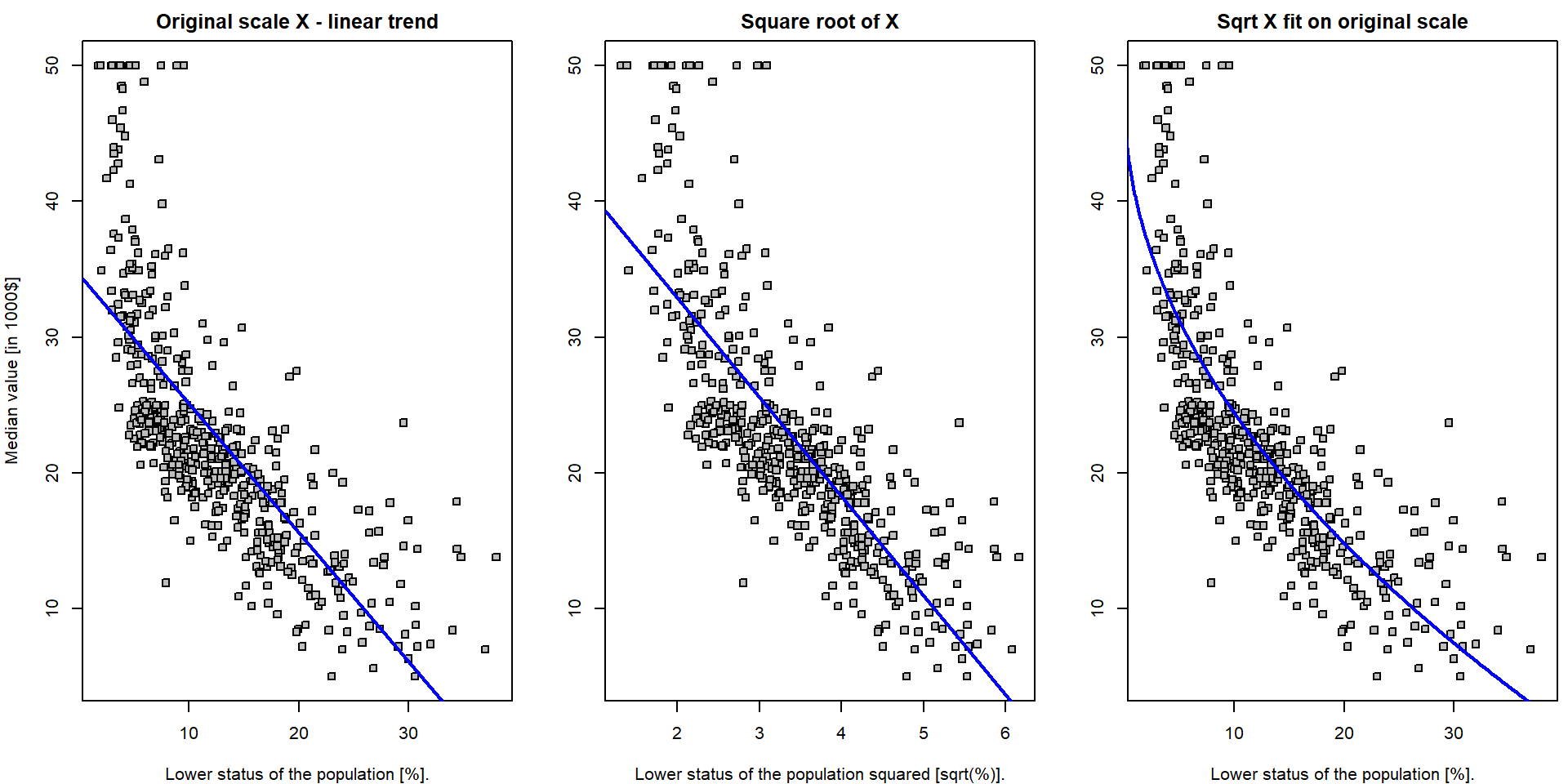
Note, that the first plot is the scatterplot for the relationship
medv \(\sim\)
lstat – which is the original model which fits the straight
line through the data. The middle plot is the scatterplot for the
relationship medv \(\sim
(\)lstat\()^{1/2}\)
and the corresponding model (lm.fit4) again fits the
straight line through the data (with coordinates \((Y_i, \sqrt{X_i})\)). Finally, the third
plot shows the original scatterplot (the data with the coordinates \((Y_i, X_i)\)) but the model
lm.fit4 does not fit a straight line through such data. The
interpretation is simple with respect to the transformed data \((Y_i, \sqrt{X_i})\) but it is not that much
straightforward with respect to the original data \((Y_i, X_i)\).
Nevertheless, it seems that the model lm.fit4 fits the
data more closely that the model lm.fit.
Individual work
- Think of some alternative transformation of the original data in order to improve the overall fit of the model.
- Do not care for the interpretation issues for now, only focus on some reasonable approximation that will help to improve the quality of the fit.
- Try to obtain some quantitative characteristic for the quality of the fits – for instance, the residual sum of squares.